4KXQ
 
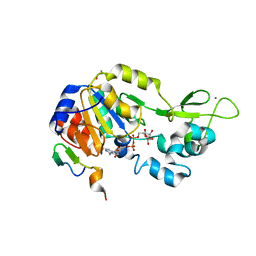 | | Structure of NAD-dependent protein deacetylase sirtuin-1 (closed state, 1.85 A) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Davenport, A.M, Huber, F.M, Hoelz, A. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structural and Functional Analysis of Human SIRT1.
J.Mol.Biol., 426, 2014
|
|
4IF5
 
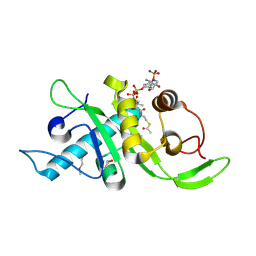 | | Structure of human Mec17 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, CHLORIDE ION | | Authors: | Davenport, A.M, Collins, L, Minor, P, Sternberg, P, Hoelz, A. | | Deposit date: | 2012-12-14 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the alpha-Tubulin Acetyltransferase MEC-17.
J.Mol.Biol., 426, 2014
|
|
4IF6
 
 | |
4IG9
 
 | |
5VDC
 
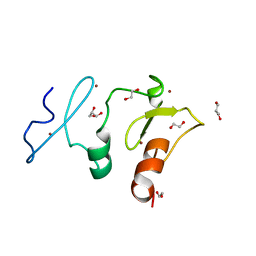 | | Crystal structure of the human DPF2 tandem PHD finger domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ZINC ION, ... | | Authors: | Huber, F.M, Davenport, A.M, Hoelz, A. | | Deposit date: | 2017-04-01 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Histone-binding of DPF2 mediates its repressive role in myeloid differentiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3IKO
 
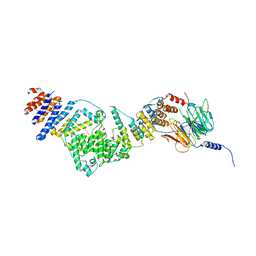 | | Crystal structure of the heterotrimeric Sec13-Nup145C-Nup84 nucleoporin complex | | Descriptor: | Nucleoporin NUP145C, Nucleoporin NUP84, Protein transport protein SEC13 | | Authors: | Nagy, V, Hsia, K.-C, Debler, E.W, Davenport, A, Blobel, G, Hoelz, A. | | Deposit date: | 2009-08-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a trimeric nucleoporin complex reveals alternate oligomerization states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3TKN
 
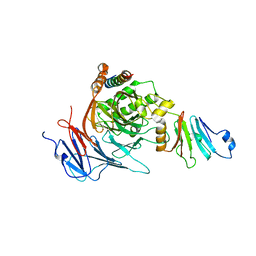 | | Structure of the Nup82-Nup159-Nup98 heterotrimer | | Descriptor: | Nucleoporin 98, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T.T, Hoelz, A. | | Deposit date: | 2011-08-28 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for the anchoring of proto-oncoprotein nup98 to the cytoplasmic face of the nuclear pore complex.
J.Mol.Biol., 419, 2012
|
|
5HB0
 
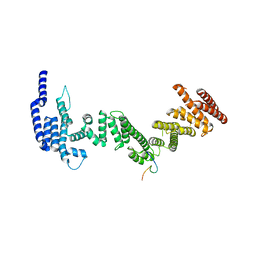 | |
5HB5
 
 | |
5HAX
 
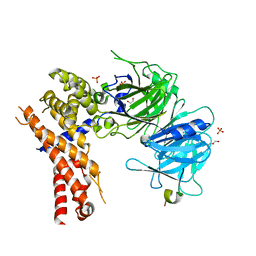 | | Crystal structure of Chaetomium thermophilum Nup170 NTD-Nup53 complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Mobbs, G, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5HB6
 
 | |
5HAY
 
 | |
5HB1
 
 | |
5HB7
 
 | |
5HB3
 
 | |
5HB2
 
 | |
5HAZ
 
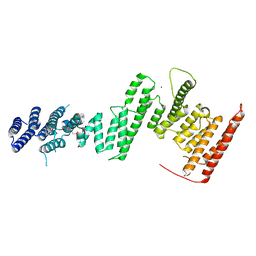 | | Structure of Chaetomium thermophilum Nup170 CTD | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5HB8
 
 | |
5HB4
 
 | |
