8SM1
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
8SM0
 
 | | Crystal structure of human complement receptor 2 (CD21) in complex with Epstein-Barr virus major glycoprotein gp350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement receptor type 2, Envelope glycoprotein gp350, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
7S0J
 
 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S08
 
 | |
7S1B
 
 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
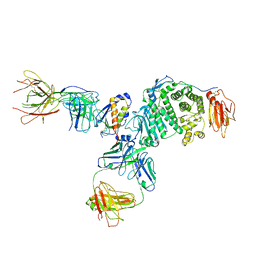 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S83
 
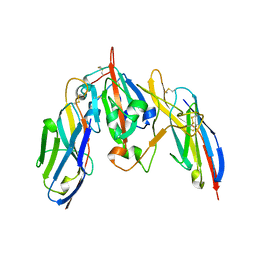 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | Authors: | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
6U6M
 
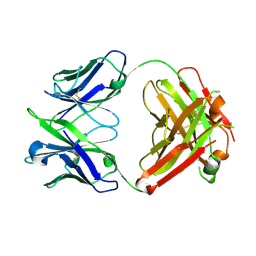 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH840.1 | | Descriptor: | DH840.1 Fab heavy chain, DH840.1 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
6U6O
 
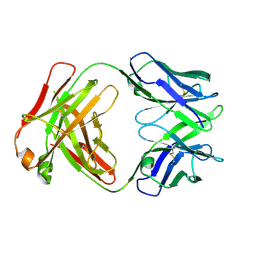 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH846 | | Descriptor: | DH846 Fab heavy chain, DH846 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
5ISX
 
 | |
5ISW
 
 | |
2ZYG
 
 | | Apo-form of dimeric 6-phosphogluconate dehydrogenase | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-21 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
2ZYD
 
 | | Dimeric 6-phosphogluconate dehydrogenase complexed with glucose | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, D-glucose | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
2ZYA
 
 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, decarboxylating | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
3FWN
 
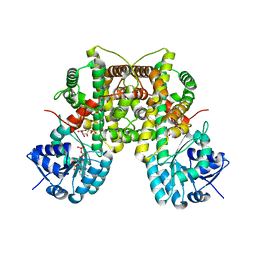 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
3PJG
 
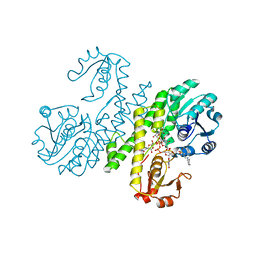 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PID
 
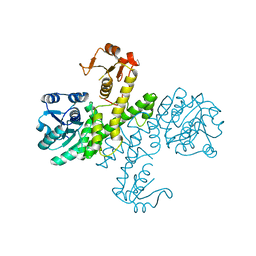 | | The apo-form UDP-glucose 6-dehydrogenase with a C-terminal six-histidine tag | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-06 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLN
 
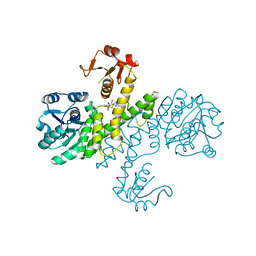 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with UDP-glucose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLR
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
