4GPS
 
 | |
4GPU
 
 | |
2ZIU
 
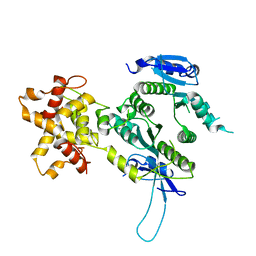 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIV
 
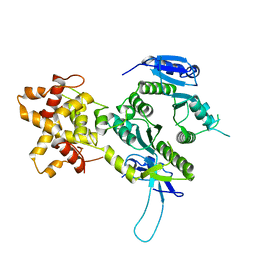 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIX
 
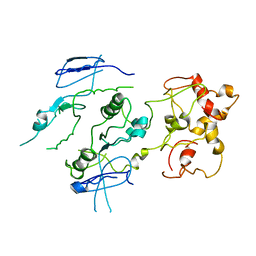 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIW
 
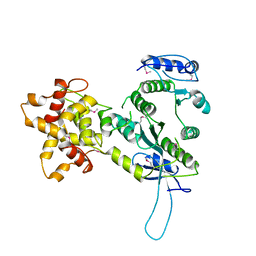 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
3PIF
 
 | |
3PIE
 
 | |
6AIX
 
 | |
6AIY
 
 | |
6IZ8
 
 | | Crystal Structure of TagF from Pseudomonas aeruginosa | | Descriptor: | Type VI secretion-associated protein | | Authors: | Chang, J.H, Ok, C.K. | | Deposit date: | 2018-12-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Type VI Secretion System Accessory Protein TagF from Pseudomonas Aeruginosa.
Protein Pept.Lett., 26, 2019
|
|
2ZU6
 
 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
4J7M
 
 | |
4J7L
 
 | |
5ELM
 
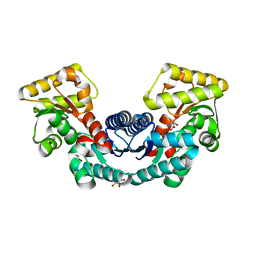 | | Crystal structure of L-aspartate/glutamate specific racemase in complex with L-glutamate | | Descriptor: | Asp/Glu_racemase family protein, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Ahn, J.W, Chang, J.H, Kim, K.J. | | Deposit date: | 2015-11-04 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an atypical active site of an l-aspartate/glutamate-specific racemase from Escherichia coli
Febs Lett., 589, 2015
|
|
5ELL
 
 | |
6KWS
 
 | |
6KWT
 
 | |
8IYI
 
 | |
4YVM
 
 | |
7BU2
 
 | | Structure of alcohol dehydrogenase YjgB from Escherichia coli | | Descriptor: | Alcohol dehydrogenase, GLYCEROL, NITRATE ION, ... | | Authors: | Nguyen, G.T, Kim, Y.-G, Ahn, J.-W, Chang, J.H. | | Deposit date: | 2020-04-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Molecules, 25, 2020
|
|
7BU3
 
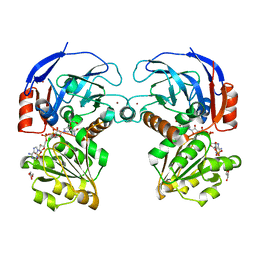 | | Structure of alcohol dehydrogenase YjgB in complex with NADP from Escherichia coli | | Descriptor: | ASPARTIC ACID, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nguyen, G.T, Kim, Y.-G, Ahn, J.-W, Chang, J.H. | | Deposit date: | 2020-04-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Molecules, 25, 2020
|
|
8I28
 
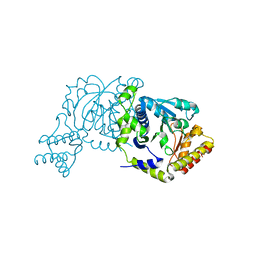 | |
6KVR
 
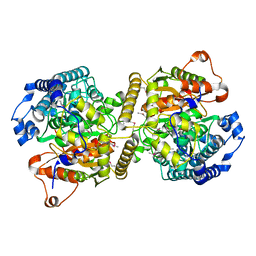 | | Fatty acid amide hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Min, C.A, Yun, J.S, Chang, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
5IM0
 
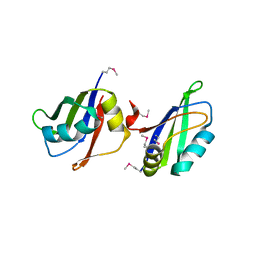 | |
