4I5M
 
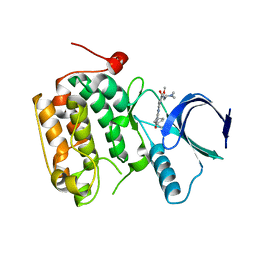 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4IFN
 
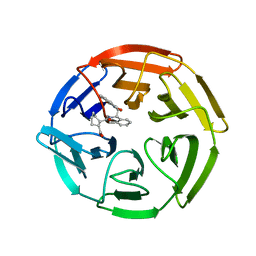 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFJ
 
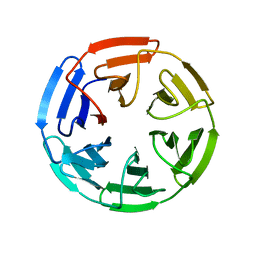 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFL
 
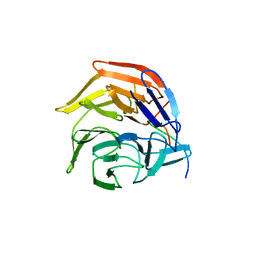 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
7KQY
 
 | | Crystal Structure and Characterization of Human Heavy-Chain only Antibodies reveals a novel, stable dimeric structure similar to Monoclonal Antibodies | | Descriptor: | Heavy-Chain only Human Antibodies | | Authors: | Bahmanjah, S, Mieczkowski, C, Yu, Y, Baker, J, Raghunathan, G, Tomazela, D, Hsieh, M, Mccoy, M, Strickland, C, Fayadat-Dilman, L. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal Structure and Characterization of Human Heavy-Chain Only Antibodies Reveals a Novel, Stable Dimeric Structure Similar to Monoclonal Antibodies.
Antibodies, 9, 2020
|
|
4Y73
 
 | | Crystal structure of IRAK4 kinase domain with inhibitor | | Descriptor: | 5-{[(1R,2S)-2-aminocyclohexyl]amino}-N-[1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Lesburg, C.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of 5-Amino-N-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide Inhibitors of IRAK4.
Acs Med.Chem.Lett., 6, 2015
|
|
5T1T
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1S
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4I5P
 
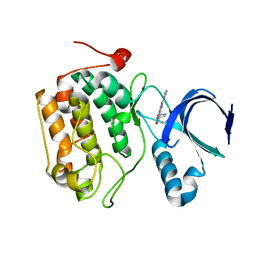 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | (7R)-8-cyclopentyl-7-ethyl-5-methyl-2-(1H-pyrrol-2-yl)-7,8-dihydropteridin-6(5H)-one, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4I6H
 
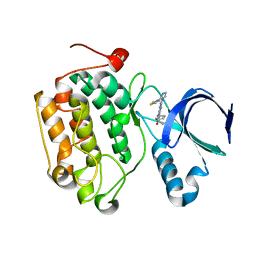 | |
4I6F
 
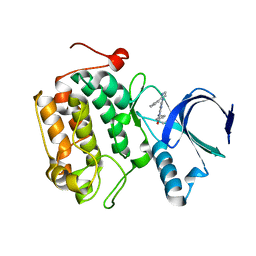 | |
4I6B
 
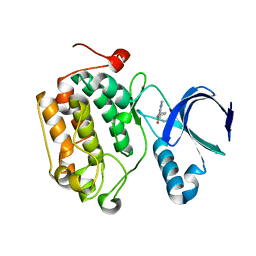 | |
4I1F
 
 | | Structure of Parkin-S223P E3 ligase | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
4I1H
 
 | | Structure of Parkin E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
8DS5
 
 | | X-ray structure of the MK5890 Fab - CD27 antibody-antigen complex | | Descriptor: | CADMIUM ION, CD27 antigen, MK-5890 Fab heavy chain, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Preclinical characterization and clinical translation of pharmacodynamic markers for MK-5890: a human CD27 activating antibody for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1O
 
 | |
7P1V
 
 | | Apo structure of KDNase from Trichophyton Rubrum | | Descriptor: | CALCIUM ION, Extracellular sialidase/neuraminidase, GLYCEROL | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1B
 
 | |
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1U
 
 | |
7P1Q
 
 | |
7P1F
 
 | | Structure of KDNase from Aspergillus terrerus in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1R
 
 | |
7P1D
 
 | |
