1MA1
 
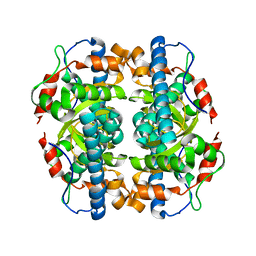 | | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Adams, J.J, Anderson, B.F, Renault, J.P, Verchere-Beaur, C, Morgenstern-Badarau, I, Jameson, G.B. | | Deposit date: | 2002-07-31 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum
To be published
|
|
2OZN
 
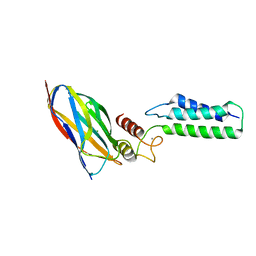 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6PRO
 
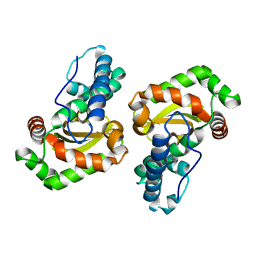 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
2AKQ
 
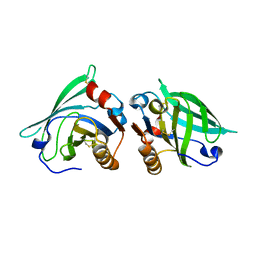 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
2B59
 
 | |
3TJH
 
 | | 42F3-p3A1/H2-Ld complex | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kruse, A, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-24 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TPU
 
 | | 42F3 p5E8/H2-Ld complex | | Descriptor: | 1,2-ETHANEDIOL, 42F3 alpha, 42F3 beta, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TFK
 
 | | 42F3-p4B10/H2-Ld | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TF7
 
 | | 42F3 QL9/H2-Ld complex | | Descriptor: | 42F3 Mut7 scFv (42F3 alpha chain, linker, 42F3 beta chain), ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3KCP
 
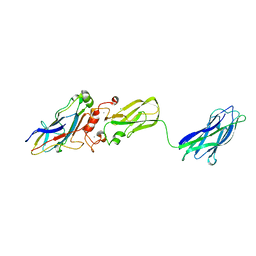 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
2VO8
 
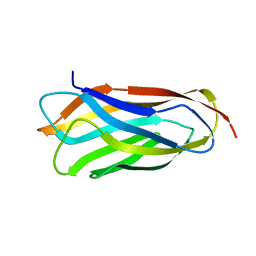 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2AEW
 
 | |
8TRV
 
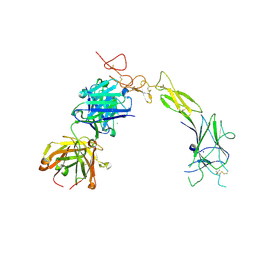 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be Published
|
|
8TV2
 
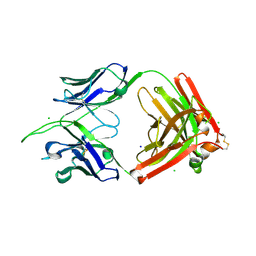 | | Structure of apo FabS1C_L1 | | Descriptor: | CHLORIDE ION, S1C variant of Fab_L1 heavy chain, S1C variant of Fab_L1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8TV1
 
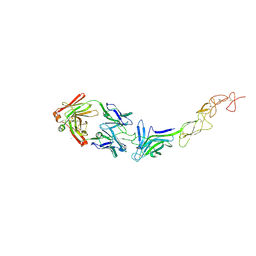 | | Structure of the EphA2 LBDCRD bound to FabS1C_L1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8TV5
 
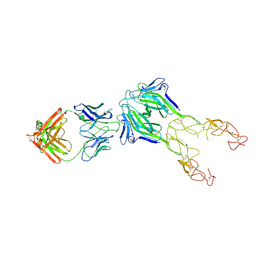 | | Structure of the EphA2 LBDCRD bound to FabS1CE_L1 in a 2:1 (EphA2 to Fab) ratio | | Descriptor: | Ephrin type-A receptor 2, MAGNESIUM ION, S1CE variant of Fab_L1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
4N0C
 
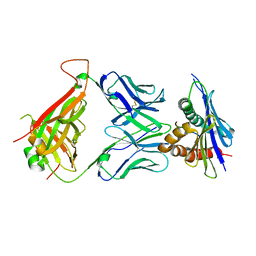 | | 42F3 TCR pCPE3/H-2Ld complex | | Descriptor: | 42F3 VmCh alpha, 42F3 VmCh beta, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-01 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4N5E
 
 | | 42F3 TCR pCPA12/H-2Ld complex | | Descriptor: | 42F3 alpha VmCh, 42F3 beta VmCh, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-09 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (3.059 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4MXQ
 
 | | 42F3 TCR pCPC5/H-2Ld Complex | | Descriptor: | 42F3 alpha VmVh chimera, 42F3 beta VmVh chimera, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-26 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4MVB
 
 | | 42F3 pCPB7/H-2Ld Complex | | Descriptor: | 42F3 alpha VmCh, 42F3 beta VmCh, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-23 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.088 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4MS8
 
 | | 42F3 TCR pCPB9/H-2Ld Complex | | Descriptor: | 42F3 alpha, 42F3 beta, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
2W1N
 
 | | cohesin and fibronectin type-III double module construct from the Clostridium perfringens glycoside hydrolase GH84C | | Descriptor: | ACETATE ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Czjzek, M, Smith, S.J, Boraston, A.B. | | Deposit date: | 2008-10-17 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
9FSE
 
 | | Human ROR2 cysteine-rich domain (CRD) and Kringle domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase transmembrane receptor ROR2 | | Authors: | Griffiths, S.C, Tan, J, Wagner, A, Blazer, L.L, Adams, J.J, Srinivasan, S, Moghisaei, S, Sidhu, S.S, Siebold, C, Ho, H.H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and function of the ROR2 cysteine-rich domain in vertebrate noncanonical WNT5A signaling.
Elife, 13, 2024
|
|
2QUG
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form A | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Hansen, G, Morton, C.J, Pearce, M.C, Feil, S.C, Adams, J.J, Parker, M.W, Bottomley, S.P. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2V5C
 
 | | Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure | | Descriptor: | CACODYLATE ION, CALCIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
