6M1V
 
 | |
2DK9
 
 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
7DEA
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
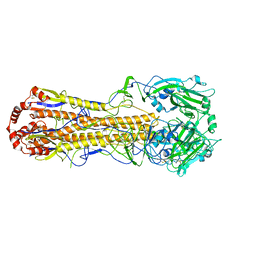 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
7CUJ
 
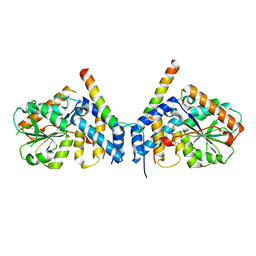 | | Crystal structure of fission yeast Ccq1 and Tpz1 | | Descriptor: | Coiled-coil quantitatively-enriched protein 1, Protection of telomeres protein tpz1 | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUH
 
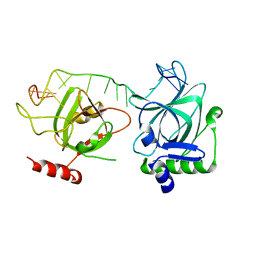 | | Crystal structure of fission yeast Pot1 and ssDNA | | Descriptor: | Protection of telomeres protein 1, Telomere single-strand DNA | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUI
 
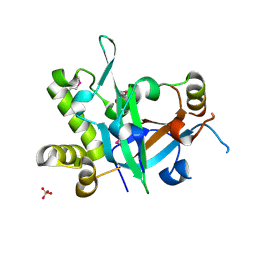 | | Crystal structure of fission yeast Pot1 and Tpz1 | | Descriptor: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7D6C
 
 | | Crystal structure of CcmM N-terminal domain in complex with CcmN | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Carboxysome assembly protein CcmN | | Authors: | Sun, H, Cui, N, Han, S.J, Chen, Z.P, Xia, L.Y, Chen, Y, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Complex structure reveals CcmM and CcmN form a heterotrimeric adaptor in beta-carboxysome.
Protein Sci., 30, 2021
|
|
1UI8
 
 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1UI7
 
 | | Site-directed mutagenesis of His433 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
5JXL
 
 | |
2UYG
 
 | |
1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
2RQ1
 
 | |
2RQ5
 
 | |
1U4Q
 
 | | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
1S35
 
 | |
1U5P
 
 | | Crystal Structure of Repeats 15 and 16 of Chicken Brain Alpha Spectrin | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Spectrin alpha chain, ... | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
