2CHY
 
 | | THREE-DIMENSIONAL STRUCTURE OF CHEY, THE RESPONSE REGULATOR OF BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Mottonen, J.M, Stock, A.M, Stock, J.B, Schutt, C.E. | | Deposit date: | 1990-05-17 | | Release date: | 1990-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis.
Nature, 337, 1989
|
|
1CHD
 
 | | CHEB METHYLESTERASE DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | West, A.H, Martinez-Hackert, E, Stock, A.M. | | Deposit date: | 1995-03-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB.
J.Mol.Biol., 250, 1995
|
|
1DZ4
 
 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ9
 
 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ6
 
 | | ferrous p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ8
 
 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
3NNN
 
 | | BeF3 Activated DrrD Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR D, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
1KGS
 
 | |
3NNS
 
 | | BeF3 Activated DrrB Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR B, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
3BS1
 
 | | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with a Novel Mode of Binding | | Descriptor: | Accessory gene regulator protein A, DNA (5'-D(*DAP*DAP*(BRU)P*DAP*DCP*DTP*DTP*DAP*DAP*DCP*DTP*DGP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DAP*DAP*DCP*DAP*DGP*DTP*DTP*DAP*DAP*DGP*(BRU)P*DAP*DT)-3'), ... | | Authors: | Sidote, D.J, Barbieri, C, Wu, T, Stock, A.M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with an Unusual Mode of Binding.
Structure, 16, 2008
|
|
1MKY
 
 | | Structural Analysis of the Domain Interactions in Der, a Switch Protein Containing Two GTPase Domains | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Probable GTP-binding protein engA | | Authors: | Robinson, V.L, Hwang, J, Fox, E, Inouye, M, Stock, A.M. | | Deposit date: | 2002-08-29 | | Release date: | 2003-01-14 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain Arrangement of Der, a Switch Protein Containing Two GTPase Domains
Structure, 10, 2002
|
|
3NHZ
 
 | | Structure of N-terminal Domain of MtrA | | Descriptor: | MAGNESIUM ION, Two component system transcriptional regulator mtrA | | Authors: | Barbieri, C.M, Mack, T.R, Robinson, V.L, Miller, M.T, Stock, A.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
1NEP
 
 | | Crystal Structure Analysis of the Bovine NPC2 (Niemann-Pick C2) Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epididymal secretory protein E1, PHOSPHATE ION | | Authors: | Friedland, N, Liou, H.-L, Lobel, P, Stock, A.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Cholesterol-binding Protein Deficient in Niemann-Pick Type C2 Disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4P7H
 
 | |
4PA0
 
 | | Omecamtiv Mercarbil binding site on the Human Beta-Cardiac Myosin Motor Domain | | Descriptor: | GLYCEROL, Myosin-7,Green fluorescent protein, methyl 4-(2-fluoro-3-{[(6-methylpyridin-3-yl)carbamoyl]amino}benzyl)piperazine-1-carboxylate | | Authors: | Winkelmann, D.A, Miller, M.T, Stock, A.M. | | Deposit date: | 2014-04-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity.
Nat Commun, 6, 2015
|
|
1OPC
 
 | |
1P2F
 
 | |
1ZGZ
 
 | |
1ZTY
 
 | |
1ZU0
 
 | |
1XHF
 
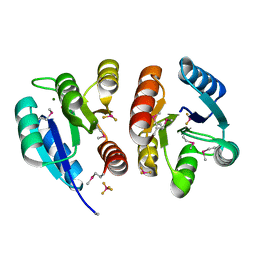 | | Crystal structure of the bef3-activated receiver domain of redox response regulator arca | | Descriptor: | Aerobic respiration control protein arcA, BERYLLIUM DIFLUORIDE, BERYLLIUM TETRAFLUORIDE ION, ... | | Authors: | Toro-Roman, A, Mack, T.R, Stock, A.M. | | Deposit date: | 2004-09-18 | | Release date: | 2005-05-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural Analysis and Solution Studies of the Activated Regulatory Domain of the Response Regulator ArcA: A Symmetric Dimer Mediated by the alpha4-beta5-alpha5 Face
J.Mol.Biol., 349, 2005
|
|
2PL9
 
 | |
2PMC
 
 | |
1XHE
 
 | |
