6QVE
 
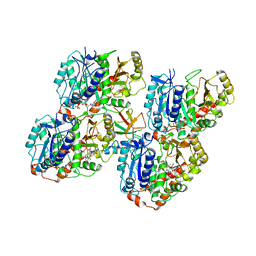 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QUY
 
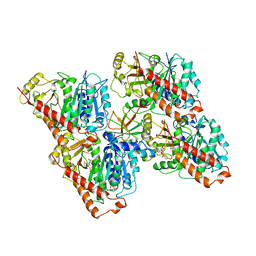 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
4TY9
 
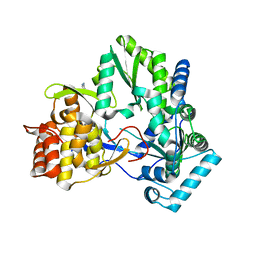 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYB
 
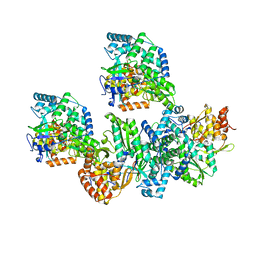 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
6QUS
 
 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
4RS4
 
 | |
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4Z44
 
 | | F454K Mutant of Tryptophan 7-halogenase PrnA | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Shepherd, S.A, Karthikeyan, C, Latham, J, Struck, A.-W, Thompson, M.L, Menon, B, Levy, C.W, Leys, D, Micklefield, J. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
2W38
 
 | | Crystal structure of the pseudaminidase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SIALIDASE | | Authors: | Xu, G, Ryan, C, Kiefel, M.J, Wilson, J.C, Taylor, G.L. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on the Pseudomonas Aeruginosa Sialidase-Like Enzyme Pa2794 Suggest Substrate and Mechanistic Variations.
J.Mol.Biol., 386, 2009
|
|
3FYS
 
 | | Crystal Structure of DegV, a fatty acid binding protein from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, PALMITIC ACID, ... | | Authors: | Nan, J, Zhou, Y.F, Yang, C. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a fatty acid-binding protein from Bacillus subtilis determined by sulfur-SAD phasing using in-house chromium radiation
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1TLB
 
 | | Yeast coproporphyrinogen oxidase | | Descriptor: | Coproporphyrinogen III oxidase, SULFATE ION | | Authors: | Phillip, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1TK1
 
 | | YEAST OXYGEN-DEPENDENT COPROPORPHYRINOGEN OXIDASE | | Descriptor: | Coproporphyrinogen III oxidase | | Authors: | Phillips, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Oxygen-dependant Coproporphyrinogen Oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
3J6Q
 
 | | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Structural protein VP3 | | Authors: | Zhu, B, Yang, C, Liu, H, Cheng, L, Song, F, Zeng, S, Huang, X, Ji, G, Zhu, P. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus.
J.Mol.Biol., 426, 2014
|
|
1TKL
 
 | | Yeast Oxygen-Dependent Coproporphyrinogen Oxidase | | Descriptor: | Coproporphyrinogen III oxidase | | Authors: | Phillips, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygen-dependant Coproporphyrinogen Oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
3OQT
 
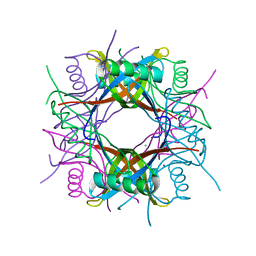 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
3ZRS
 
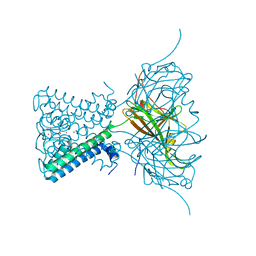 | | X-ray crystal structure of a KirBac potassium channel highlights a mechanism of channel opening at the bundle-crossing gate. | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION | | Authors: | Bavro, V.N, De Zorzi, R, Schmidt, M.R, Muniz, J.R.C, Zubcevic, L, Sansom, M.S.P, Venien-Bryan, C, Tucker, S.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a Kirbac Potassium Channel with an Open Bundle Crossing Indicates a Mechanism of Channel Gating
Nat.Struct.Mol.Biol., 19, 2012
|
|
1I7Q
 
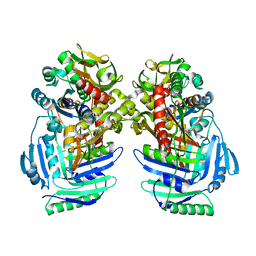 | | ANTHRANILATE SYNTHASE FROM S. MARCESCENS | | Descriptor: | ANTHRANILATE SYNTHASE, BENZOIC ACID, GLUTAMIC ACID, ... | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I7S
 
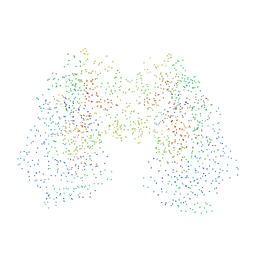 | | ANTHRANILATE SYNTHASE FROM SERRATIA MARCESCENS IN COMPLEX WITH ITS END PRODUCT INHIBITOR L-TRYPTOPHAN | | Descriptor: | ANTHRANILATE SYNTHASE, TRPG, TRYPTOPHAN | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3CE7
 
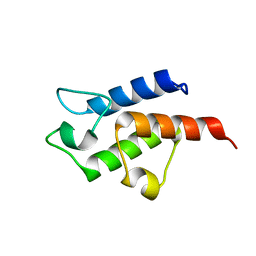 | | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510 | | Descriptor: | Specific mitochondrial acyl carrier protein | | Authors: | Wernimont, A.K, Dong, A, Yang, C, Khuu, C, Lam, A, Brand, V, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510.
To be Published
|
|
4K7E
 
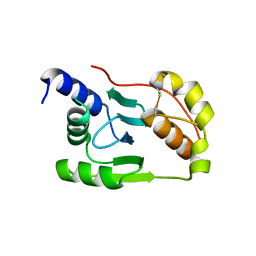 | | Crystal structure of Junin virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Zhang, Y.J, Li, L, Liu, X, Dong, S.S, Wang, W.M, Huo, T, Rao, Z.H, Yang, C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Junin virus nucleoprotein
J.Gen.Virol., 94, 2013
|
|
7ADI
 
 | | KirBac3.1 W46R: role of a highly conserved tryptophan at the membrane-water interface of Kir channel | | Descriptor: | Inward rectifier potassium channel Kirbac3.1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Venien-Bryan, C, Fagnen, C, De Zorzi, R, Bannwarth, L, Oubella, I, Haouz, A. | | Deposit date: | 2020-09-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Integrative Study of the Structural and Dynamical Properties of a KirBac3.1 Mutant: Functional Implication of a Highly Conserved Tryptophan in the Transmembrane Domain.
Int J Mol Sci, 23, 2021
|
|
7ZDZ
 
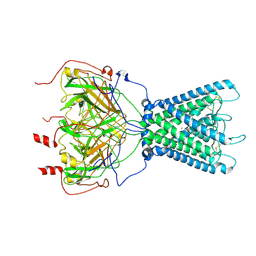 | | Cryo-EM structure of the human inward-rectifier potassium 2.1 channel (Kir2.1) | | Descriptor: | Inward rectifier potassium channel 2, POTASSIUM ION, STRONTIUM ION | | Authors: | Fernandes, C.A.H, Venien-Bryan, C, Fagnen, C, Zuniga, D. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy unveils unique structural features of the human Kir2.1 channel.
Sci Adv, 8, 2022
|
|
