6H75
 
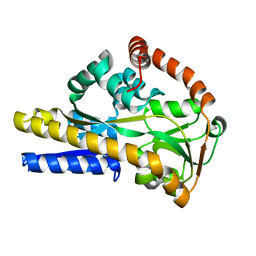 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
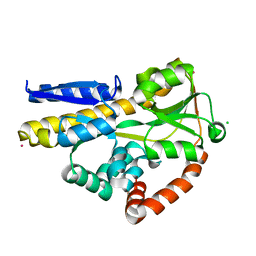 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3D55
 
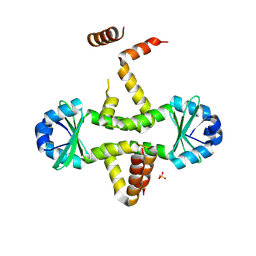 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
5FWN
 
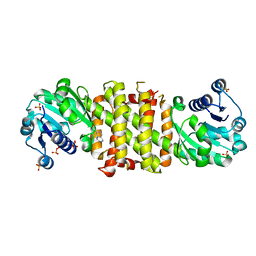 | | Imine Reductase from Amycolatopsis orientalis. Closed form in in complex with (R)- Methyltetrahydroisoquinoline | | Descriptor: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
3CAL
 
 | |
6S5Z
 
 | | Structure of Rib R28N from Streptococcus pyogenes | | Descriptor: | SODIUM ION, Surface protein R28 | | Authors: | Whelan, F, Griffiths, S.C, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5W
 
 | | Structure of Rib domain 'Rib Long' from Lactobacillus acidophilus | | Descriptor: | SODIUM ION, SULFATE ION, Surface protein | | Authors: | Griffiths, S.C, Cooper, R.E.M, Whelan, F, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5Y
 
 | |
6T9M
 
 | | Crystal structure of the Chitinase Domain of the Spore Coat Protein CotE from Clostridium difficile | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptide in active site, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2RKZ
 
 | |
2RL0
 
 | |
2RKY
 
 | |
6FF3
 
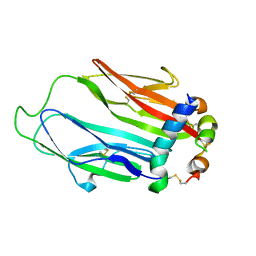 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L1 with Human IGF-I | | Descriptor: | Insulin-like growth factor I, Neural/ectodermal development factor IMP-L2 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D, Viola, C.M. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
6FEY
 
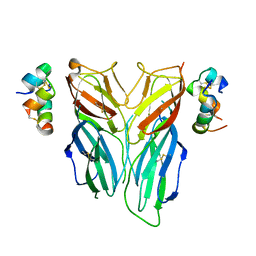 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L2 with Drosophila DILP5 insulin | | Descriptor: | Neural/ectodermal development factor IMP-L2, Probable insulin-like peptide 5 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
6G1H
 
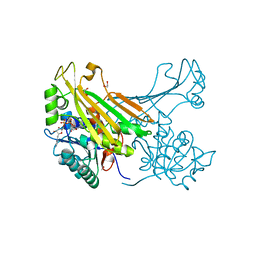 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6G1M
 
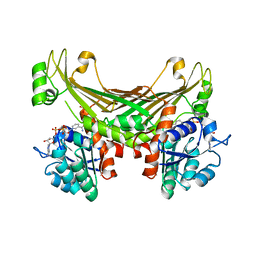 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
1GQZ
 
 | |
2W1W
 
 | | Native structure of a family 35 carbohydrate binding module from Clostridium thermocellum | | Descriptor: | CALCIUM ION, GLYCEROL, LIPOLYTIC ENZYME, ... | | Authors: | Gloster, T.M, Davies, G.J, Correia, M, Prates, J, Fontes, C, Gilbert, H.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6ZPY
 
 | | Structure of Arabinose-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPS
 
 | |
6ZPX
 
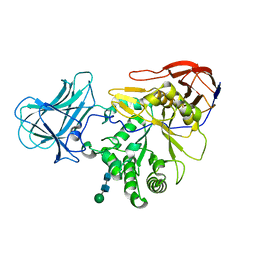 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPW
 
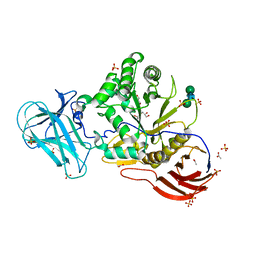 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPZ
 
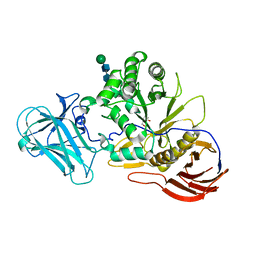 | | Structure of a-l-AraCS-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2VZQ
 
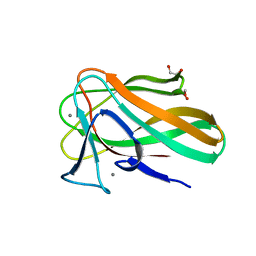 | |
2VZP
 
 | |
