1DAK
 
 | | DETHIOBIOTIN SYNTHETASE FROM ESCHERICHIA COLI, COMPLEX REACTION INTERMEDIATE ADP AND MIXED ANHYDRIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DETHIOBIOTIN SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1E3I
 
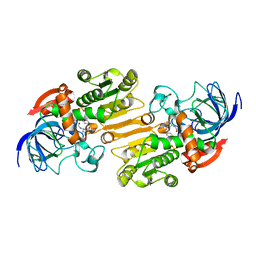 | | Mouse class II alcohol dehydrogenase complex with NADH and inhibitor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-16 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3E
 
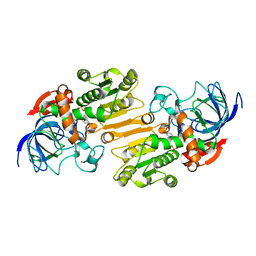 | | Mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoeoeg, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-14 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3L
 
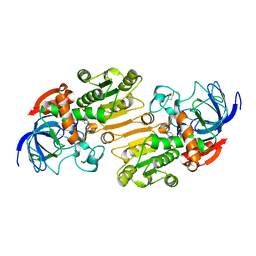 | | P47H mutant of mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | ALCOHOL DEHYDROGENASE, CLASS II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-19 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E5Q
 
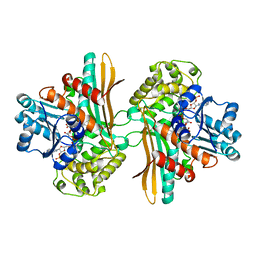 | | Ternary complex of saccharopine reductase from Magnaporthe grisea, NADPH and saccharopine | | Descriptor: | N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase [NADP(+), ... | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-28 | | Release date: | 2000-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
1H7W
 
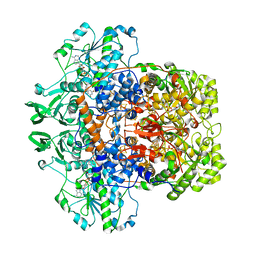 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
1H75
 
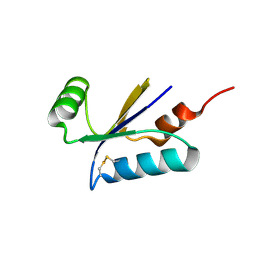 | | Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like protein NrdH-redoxin from Escherichia coli. | | Descriptor: | GLUTAREDOXIN-LIKE PROTEIN NRDH | | Authors: | Stehr, M, Schneider, G, Aslund, F, Holmgren, A, Lindqvist, Y. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-09 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Thioredoxin-Like Activity Profile of the Glutaredoxin-Like Nrdh-Redoxin from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1GV9
 
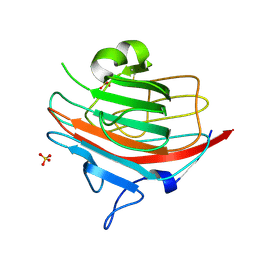 | | p58/ERGIC-53 | | Descriptor: | P58/ERGIC-53, SULFATE ION | | Authors: | Velloso, L.M, Svensson, K, Schneider, G, Pettersson, R.F, Lindqvist, Y. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-28 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of P58/Ergic-53, a Protein Involved in Glycoprotein Export from the Endoplasmic Reticulum.
J.Biol.Chem., 277, 2002
|
|
4AN4
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, GLYCOSYL TRANSFERASE | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-15 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4BB0
 
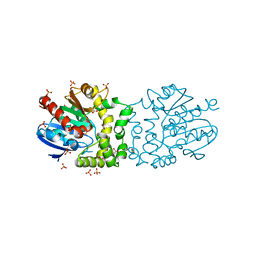 | |
4B9E
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BAZ
 
 | |
4BKL
 
 | | Crystal structure of the arthritogenic antibody M2139 (Fab fragment) in complex with the triple-helical J1 peptide | | Descriptor: | J1 EPITOPE, M2139 FAB FRAGMENT HEAVY CHAIN, M2139 FAB FRAGMENT LIGHT CHAIN | | Authors: | Raposo, B, Dobritzsch, D, Ge, C, Ekman, D, Lindh, I, Foerster, M, Uysal, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2013-04-26 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Epitope-Specific Antibody Response is Controlled by Immunoglobulin Vh Polymorphisms.
J.Exp.Med., 211, 2014
|
|
4BAU
 
 | | Structure of a putative epoxide hydrolase t131d mutant from Pseudomonas aeruginosa, with bound MFA | | Descriptor: | CHLORIDE ION, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Putative Epoxide Hydrolase T131D Mutant from Pseudomonas Aeruginosa, with Bound Mfa
To be Published
|
|
4B9A
 
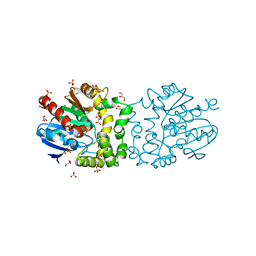 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BAT
 
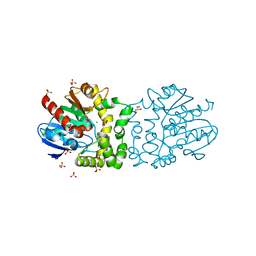 | |
4BNU
 
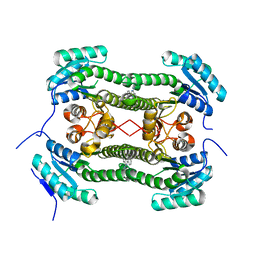 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-phenyl-4-(1,2,4- triazol-4-yl)quinazoline at 2.0A resolution | | Descriptor: | 2-phenyl-4-(1,2,4-triazol-4-yl)quinazoline, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO4
 
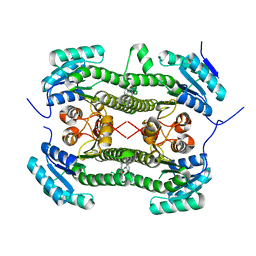 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-methoxyphenyl)-3,4- dihydro-2H-quinoline-1-carboxamide at 2.7A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-methoxyphenyl)-3,4-dihydro-2H-quinoline-1-carboxamide | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO2
 
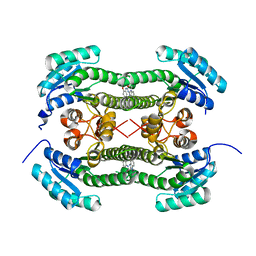 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(1-ethylbenzimidazol-2- yl)-3-(2-methoxyphenyl)urea at 1.9A resolution | | Descriptor: | 1-(1-ethylbenzimidazol-2-yl)-3-(2-methoxyphenyl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNV
 
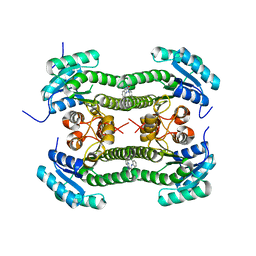 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(2-chlorophenyl)-3-(1- methylbenzimidazol-2-yl)urea at 2.5A resolution | | Descriptor: | 1-(2-chlorophenyl)-3-(1-methylbenzimidazol-2-yl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in Resolution
Acs Chem.Biol., 8, 2013
|
|
4BO8
 
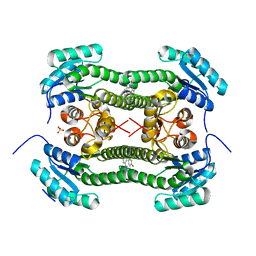 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(2-amino-4- phenylimidazol-1-yl)-3-(2-fluorophenyl)urea at 2.7A resolution | | Descriptor: | 1-(2-AMINO-4-PHENYLIMIDAZOL-1-YL)-3-(2-FLUOROPHENYL)UREA, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, SULFATE ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNX
 
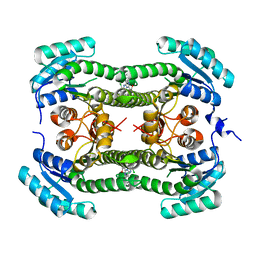 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 6-(4-(2-chloroanilino)- 1H-quinazolin-2-ylidene)cyclohexa-2, 4-dien-1-one at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 6-[4-(2-chloroanilino)-1H-quinazolin-2-ylidene]cyclohexa-2,4-dien-1-one | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO9
 
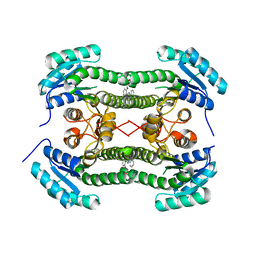 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 5-(2-(furan-2-ylmethoxy) phenyl)-2-phenyltetrazole at 2.9A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 5-[2-(FURAN-2-YLMETHOXY)PHENYL]-2-PHENYLTETRAZOLE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO1
 
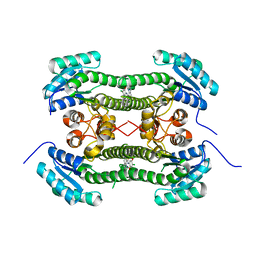 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(4-chloro-2,5- dimethoxyphenyl)quinoline-8-carboxamide at 2.2A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(4-CHLORO-2,5-DIMETHOXYPHENYL)QUINOLINE-8-CARBOXAMIDE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNW
 
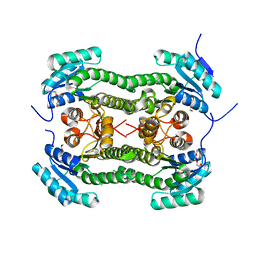 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with an unknown ligand at 1. 6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
