4OQJ
 
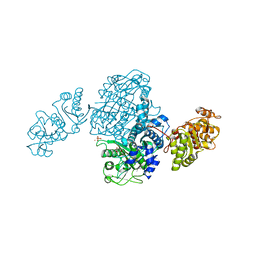 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OAT
 
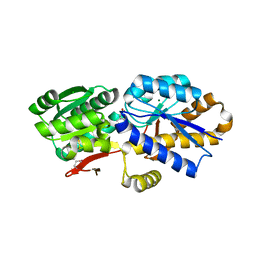 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with isoleucine. | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-06 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with isoleucine.
To be Published
|
|
3CI6
 
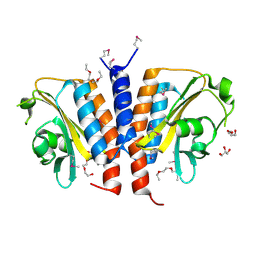 | | Crystal structure of the GAF domain from Acinetobacter phosphoenolpyruvate-protein phosphotransferase | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cuff, M.E, Shackelford, G, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the GAF domain from Acinetobacter phosphoenolpyruvate-protein phosphotransferase.
TO BE PUBLISHED
|
|
3D3O
 
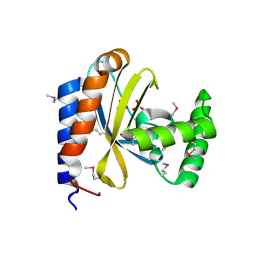 | |
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OAD
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Niedzialkowska, E, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol
To be Published
|
|
4W66
 
 | |
4OVY
 
 | | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Chang, C, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776
To be published
|
|
4OG2
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine
To be Published
|
|
4OTZ
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CYSTEINE, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein
To be Published
|
|
4OBB
 
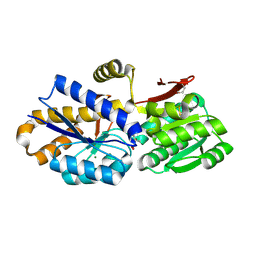 | | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid. | | Descriptor: | (3S)-3-methyl-2-oxopentanoic acid, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid.
To be Published
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4OVX
 
 | | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase domain protein TIM barrel | | Authors: | Chang, C, Bigelow, L, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-22 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776
To be published
|
|
4G6Q
 
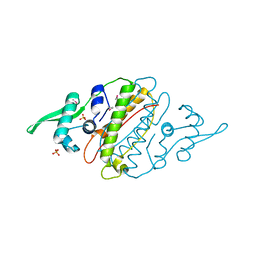 | | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Tan, K, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836, CASP Target
To be Published
|
|
3V77
 
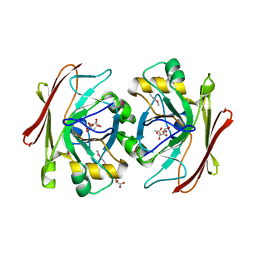 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | Descriptor: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
4GMD
 
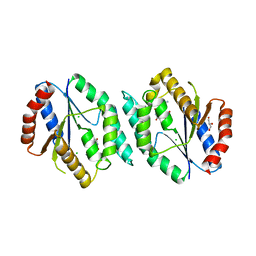 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate
To be Published
|
|
3UK0
 
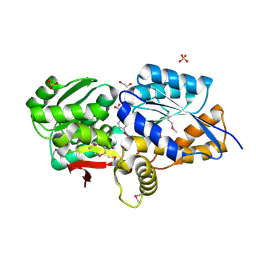 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4OVK
 
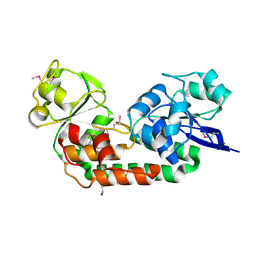 | |
3D3Y
 
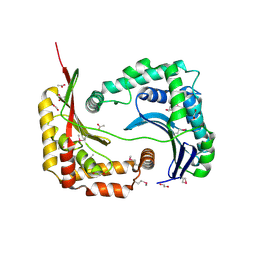 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-13 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
3UKJ
 
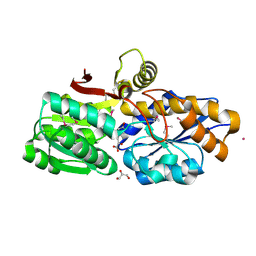 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3V7B
 
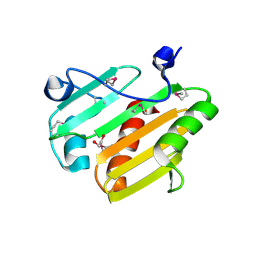 | | Dip2269 protein from corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein | | Authors: | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
4GAK
 
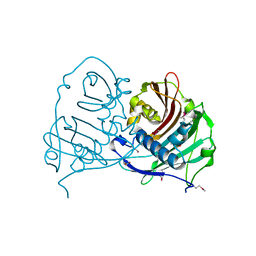 | | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale | | Descriptor: | Acyl-ACP thioesterase, CHLORIDE ION, GLYCEROL | | Authors: | Chang, C, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of acyl-ACP thioesterase from Spirosoma linguale
To be Published
|
|
3CNI
 
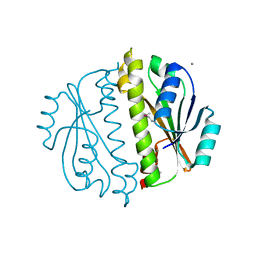 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
3CEX
 
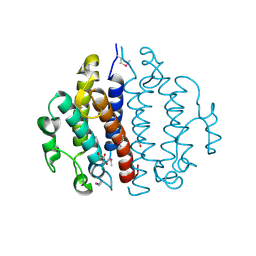 | | Crystal structure of the conserved protein of locus EF_3021 from Enterococcus faecalis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Cuff, M.E, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the conserved protein of locus EF_3021 from Enterococcus faecalis.
TO BE PUBLISHED
|
|
3CZP
 
 | | Crystal structure of putative polyphosphate kinase 2 from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Evdokimova, E, Osipiuk, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
