5W7D
 
 | | Murine acyloxyacyl hydrolase (AOAH), S262A mutant | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W7J
 
 | | X-ray structure of the E89A variant of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
1AKT
 
 | | G61N OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
6LCW
 
 | | Crosslinked alpha(Ni)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KPG
 
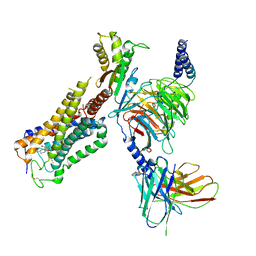 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
7CJ4
 
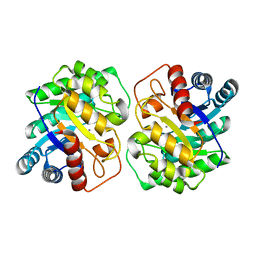 | |
5WBF
 
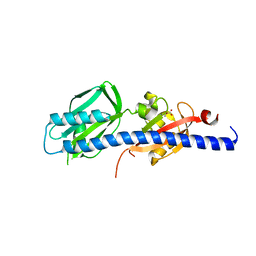 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
7CJ9
 
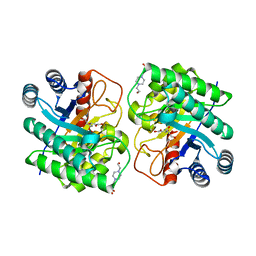 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. with additional C-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-fructose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ8
 
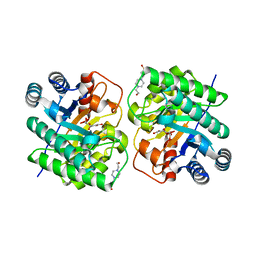 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. in complex with D-allulose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-psicose, Epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ6
 
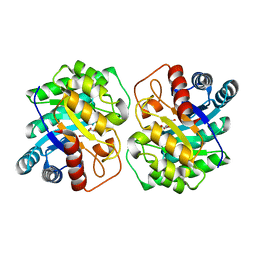 | |
7CJ7
 
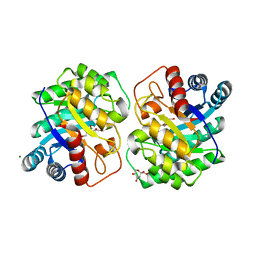 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with L-tagatose | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)oxane-2,3,4,5-tetrol, Epimerase, L-sorbose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
1ABQ
 
 | |
7CJ5
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with D-fructose | | Descriptor: | D-fructose, Epimerase, MAGNESIUM ION, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
5WMV
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
1A6S
 
 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
1G6X
 
 | | ULTRA HIGH RESOLUTION STRUCTURE OF BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) MUTANT WITH ALTERED BINDING LOOP SEQUENCE | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Addlagatta, A, Czapinska, H, Krzywda, S, Otlewski, J, Jaskolski, M. | | Deposit date: | 2000-11-08 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Ultrahigh-resolution structure of a BPTI mutant.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
2PDG
 
 | | Human aldose reductase with uracil-type inhibitor at 1.42A. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {3-[(5-CHLORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
7C3T
 
 | | Crystal structure of NE0047 (N66Q) mutant in complex with 8-azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
3NP1
 
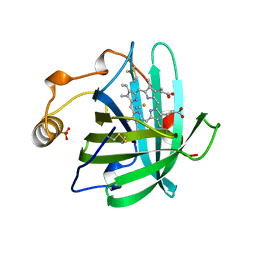 | | CRYSTAL STRUCTURE OF THE COMPLEX OF NITROPHORIN 1 FROM RHODNIUS PROLIXUS WITH CYANIDE | | Descriptor: | CYANIDE ION, NITROPHORIN 1, PHOSPHATE ION, ... | | Authors: | Weichsel, A, Andersen, J.F, Champagne, D.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a nitric oxide transport protein from a blood-sucking insect.
Nat.Struct.Biol., 5, 1998
|
|
7C3U
 
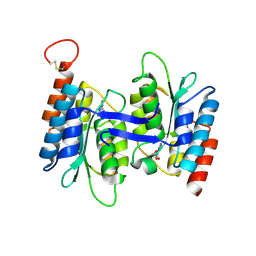 | | Crystal structure of NE0047 (N66A) mutant in complex with 8-azaguanine | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
1BS6
 
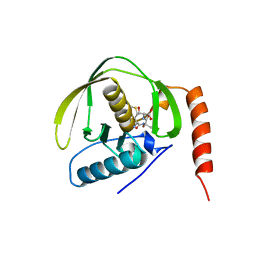 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
