3MO5
 
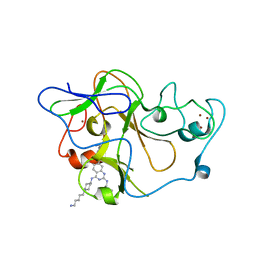 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E72 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-[1-(5-aminopentyl)piperidin-4-yl]-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
5KE7
 
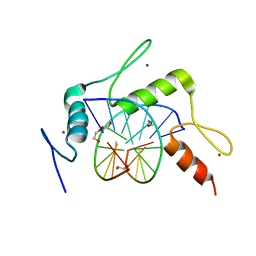 | | mouse Klf4 ZnF1-3 and TpG/MpA sequence DNA complex structure | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*AP*CP*CP*TP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
8GXP
 
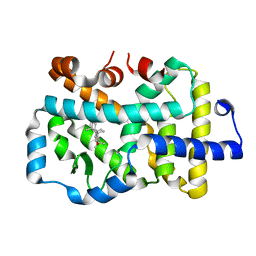 | | Complex structure of RORgama with betulinic acid | | Descriptor: | Betulinic acid, Nuclear receptor ROR-gamma | | Authors: | Zhang, X.L, Xu, C, Bai, F. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery, structural optimization, and anti-tumor bioactivity evaluations of betulinic acid derivatives as a new type of ROR gamma antagonists.
Eur.J.Med.Chem., 257, 2023
|
|
5KE6
 
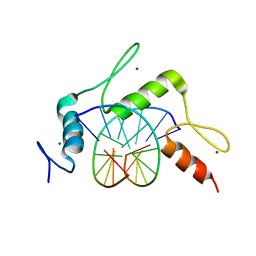 | | mouse Klf4 ZnF1-3 and TpG/CpA sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*AP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5KEB
 
 | |
5K5I
 
 | |
2HDZ
 
 | | Crystal Structure Analysis of the UBF HMG box5 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Rong, H, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-06-21 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human upstream binding factor HMG box 5 and site for binding of the cell-cycle regulatory factor TAF1
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HNB
 
 | | Solution Structure of a bacterial holo-flavodoxin | | Descriptor: | Protein mioC | | Authors: | Hu, Y, Jin, C. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
7VOA
 
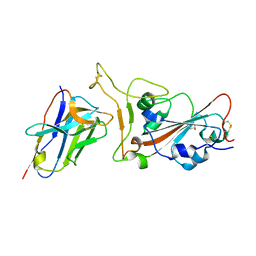 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
5K5L
 
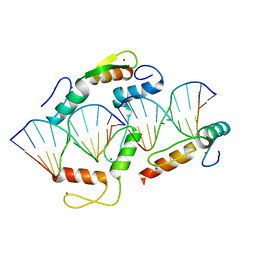 | |
5K5H
 
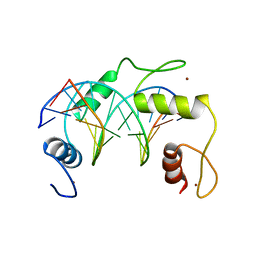 | | Homo sapiens CCCTC-binding factor (CTCF) ZnF4-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-05-23 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5KE8
 
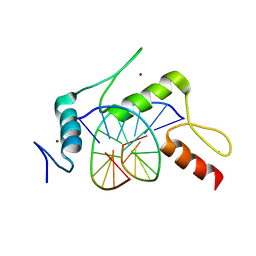 | | mouse Klf4 E446P ZnF1-3 and MpG/MpG sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5K5J
 
 | |
5HQC
 
 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5B6K
 
 | |
1T34
 
 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2004-04-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
2JSO
 
 | | Antimicrobial resistance protein | | Descriptor: | Polymyxin resistance protein pmrD | | Authors: | Jin, C, Fu, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
5HQB
 
 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
 | |
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
4PBA
 
 | |
5HQ4
 
 | | A Glycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
