2HXP
 
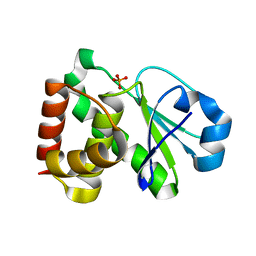 | | Crystal Structure of the human phosphatase (DUSP9) | | Descriptor: | Dual specificity protein phosphatase 9, PHOSPHATE ION | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2OOF
 
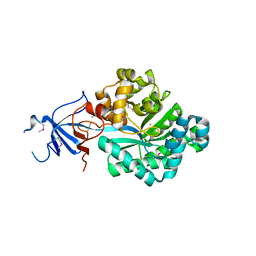 | |
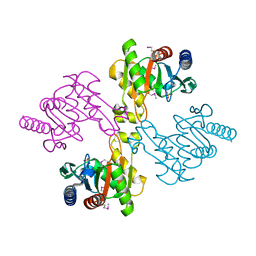
2PB9
 
 | |
2PBE
 
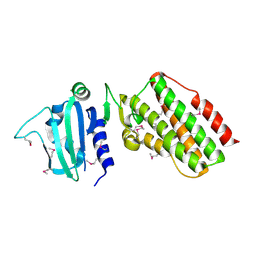 | |
2QQ6
 
 | |
4NEC
 
 | | Conversion of a Disulfide Bond into a Thioacetal Group during Echinomycin Biosynthesis | | Descriptor: | 2-CARBOXYQUINOXALINE, ACETATE ION, Echinomycin, ... | | Authors: | Hotta, K, Keegan, R.M, Ranganathan, S, Fang, M, Bibby, J, Winn, M.D, Sato, M, Lian, M, Watanabe, K, Rigden, D.J, Kim, C.-Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conversion of a disulfide bond into a thioacetal group during echinomycin biosynthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
2POF
 
 | |
2RK9
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01 | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Tyagi, R, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01.
To be Published
|
|
2PLG
 
 | |
2POZ
 
 | |
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2QVG
 
 | |
2PHP
 
 | |
2OUX
 
 | |
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
3C4R
 
 | | Crystal structure of an uncharacterized protein encoded by cryptic prophage | | Descriptor: | Uncharacterized protein | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, Ozyurt, S, Luz, J, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-19 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized protein encoded by cryptic prophage.
To be Published
|
|
2PUZ
 
 | | Crystal structure of Imidazolonepropionase from Agrobacterium tumefaciens with bound product N-formimino-L-Glutamate | | Descriptor: | CHLORIDE ION, FE (III) ION, Imidazolonepropionase, ... | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
2IOJ
 
 | |
2F1R
 
 | | Crystal Structure of molybdopterin-guanine biosynthesis protein B (mobB) | | Descriptor: | CHLORIDE ION, PRASEODYMIUM ION, molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) | | Authors: | Damodharan, L, Eswaramoorthy, S, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of molybdopterin-guanine dinucleotide biosynthesis protein B (mobB)
To be Published
|
|
1RTT
 
 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | Descriptor: | SULFATE ION, conserved hypothetical protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2GVC
 
 | | Crystal structure of flavin-containing monooxygenase (FMO)from S.pombe and substrate (methimazole) complex | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2N3O
 
 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
1RRM
 
 | |
1RVH
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER GCAAAATTTTGC | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*AP*TP*TP*TP*TP*GP*C)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
