6KZO
 
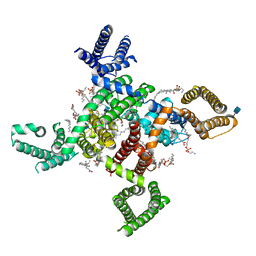 | | membrane protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6LSF
 
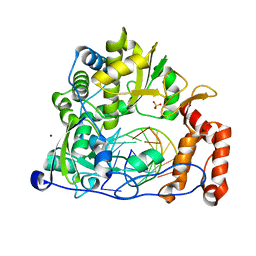 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6RA/C2S6RB form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6LSE
 
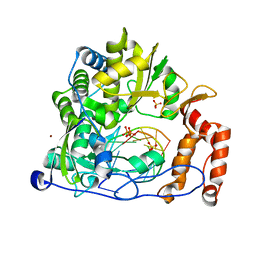 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C3S6A/C3S6B form) | | Descriptor: | Genome polyprotein, PYROPHOSPHATE 2-, RNA (35-MER), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
5YGF
 
 | |
5YGD
 
 | |
5YJO
 
 | |
5YGB
 
 | |
5YGC
 
 | |
7CC2
 
 | |
7DT2
 
 | |
7E5C
 
 | | Bacterial prolidase mutant D45W/L225Y/H226L/H343I | | Descriptor: | MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Yang, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis
Catalysis Science And Technology, 11, 2021
|
|
7DTE
 
 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
1MDR
 
 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
1MNS
 
 | |
1NX8
 
 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
1NX4
 
 | | The crystal structure of carbapenem synthase (CarC) | | Descriptor: | 2-OXOGLUTARIC ACID, Carbapenem synthase, FE (III) ION | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-08 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
3NBJ
 
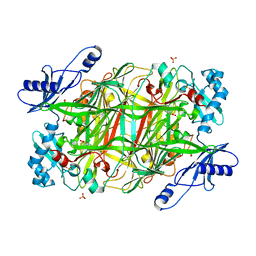 | | Crystal Structure of Y305F mutant of the copper amine oxidase from Hansenula polymorpha expressed in yeast | | Descriptor: | COPPER (II) ION, PHOSPHATE ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3N9H
 
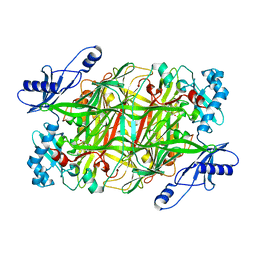 | | Crystal Structural of mutant Y305A in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-05-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3NBB
 
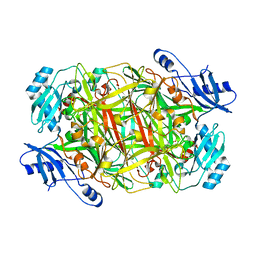 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
