7BB4
 
 | | Crystal structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose, ... | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBI
 
 | | Joint X-ray/neutron room temperature structure of H/D-exchanged PLL lectin | | Descriptor: | PLL lectin | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBC
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated PLL lectin in complex with perdeuterated L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.84 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
8I7E
 
 | | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Kumar, N, Dilawari, R, Chaubey, G.K, Modanwal, R, Talukdar, S, Dhiman, A, Chaudhary, S, Patidar, A, Kumar, A, Raje, C.I, Raje, M, Kumaran, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A
To Be Published
|
|
6JBP
 
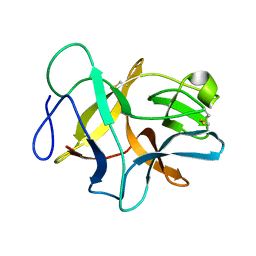 | | Structure of MP-4 from Mucuna pruriens at 2.22 Angstroms | | Descriptor: | Kunitz-type trypsin inhibitor-like 2 protein | | Authors: | Jain, A, Shikhi, M, Kumar, A, Kumar, A, Nair, D.T, Salunke, D.M. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | The structure of MP-4 from Mucuna pruriens at 2.22 angstrom resolution.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
9BDE
 
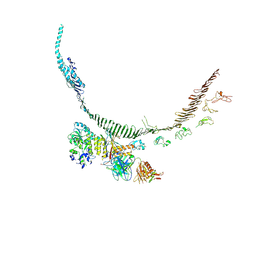 | | Middle Region of Apolipoprotein B 100 bound to Low Density Lipoprotein Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, Apolipoprotein B 100, ... | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BD8
 
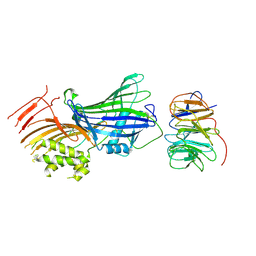 | | ApoB 100 beta barrel bound to LDLR beta propeller | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apolipoprotein B-100, Low-density lipoprotein receptor | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BD1
 
 | | beta/alpha1 region of ApoB 100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apolipoprotein B-100 | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BDT
 
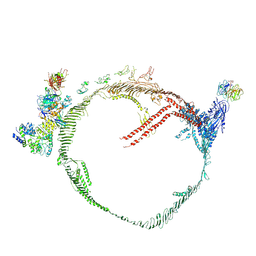 | | Apolipoprotein B 100 bound to LDL receptor and legobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, ... | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-12 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
5XYL
 
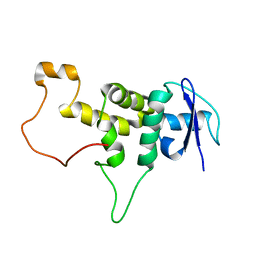 | |
9IIL
 
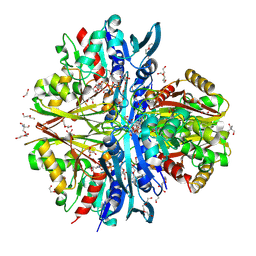 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution
To Be Published
|
|
9IIM
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution.
To Be Published
|
|
9IJ6
 
 | | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution.
To Be Published
|
|
8KDZ
 
 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-Adenosyl-L-homocysteine and Caffeic acid phenethyl ester | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase | | Authors: | Bhutkar, M, Kumar, A, Tomar, S, Kumar, P. | | Deposit date: | 2023-08-10 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering Antiviral Mechanisms of Herbacetin and Caffeic acid phenethyl ester against Chikungunya and Dengue virus, with insights into Dengue methyltransferase-CAPE crystal structure
To Be Published
|
|
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGP
 
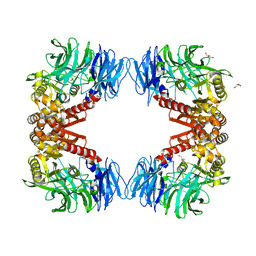 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGQ
 
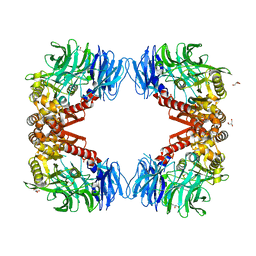 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
7F8S
 
 | | Pennisetum glaucum (Pearl millet) dehydroascorbate reductase (DHAR) with catalytic cysteine (Cy20) in sulphenic and sulfinic acid forms. | | Descriptor: | Dehydroascorbate reductase, SULFATE ION | | Authors: | Das, B.K, Kumar, A, Sreeshma, N.S, Arockiasamy, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Biochem.Biophys.Res.Commun., 591, 2021
|
|
8ZN1
 
 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
8ZOZ
 
 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
6KP1
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-methionine amino acid | | Descriptor: | METHIONINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KP0
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-arginine | | Descriptor: | ARGININE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOZ
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | Descriptor: | LEUCINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOY
 
 | | Crystal structure of two domain M1 Zinc metallopeptidase E323A mutant bound to L-tryptophan amino acid | | Descriptor: | TRYPTOPHAN, ZINC ION, Zinc metalloprotease | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
