2Y7D
 
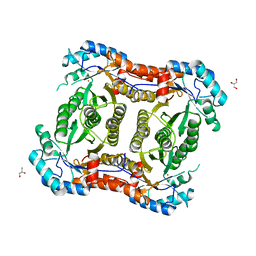 | |
1IS9
 
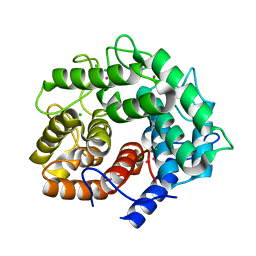 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JHL
 
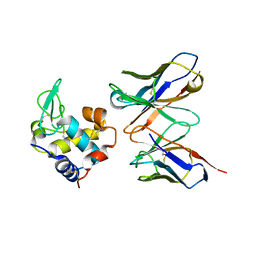 | | THREE-DIMENSIONAL STRUCTURE OF A HETEROCLITIC ANTIGEN-ANTIBODY CROSS-REACTION COMPLEX | | Descriptor: | IGG1-KAPPA D11.15 FV (HEAVY CHAIN), IGG1-KAPPA D11.15 FV (LIGHT CHAIN), PHEASANT EGG WHITE LYSOZYME | | Authors: | Chitarra, V, Alzari, P.M, Bentley, G.A, Bhat, T.N, Eisele, J.-L, Poljak, R.J. | | Deposit date: | 1993-05-04 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1L1Y
 
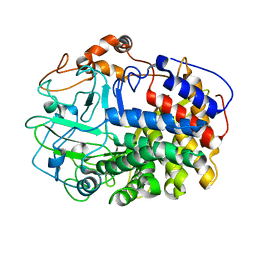 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L2A
 
 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1MZ5
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
1MS1
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MR5
 
 | | Orthorhombic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS8
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS5
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase, soaked with N-acetylneuraminyl-a-2,3-thio-galactoside (NA-S-Gal) | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MZ6
 
 | | Trypanosoma rangeli sialidase in complex with the inhibitor DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
1MS0
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA)and lactose | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS3
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS4
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS9
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme of Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1JTG
 
 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
4UHT
 
 | |
4UHK
 
 | |
4UHS
 
 | |
4UHJ
 
 | |
1W62
 
 | |
1A4E
 
 | | CATALASE A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | AZIDE ION, CATALASE A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mate, M.J. | | Deposit date: | 1998-01-29 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of catalase-A from Saccharomyces cerevisiae.
J.Mol.Biol., 286, 1999
|
|
8P5W
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5S
 
 | | Crystal structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Boyko, A, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5U
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
