7FAW
 
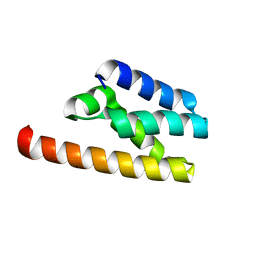 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
7FAX
 
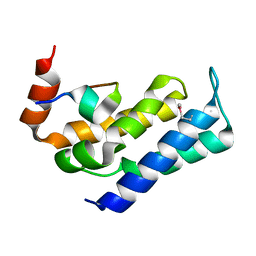 | |
8XN7
 
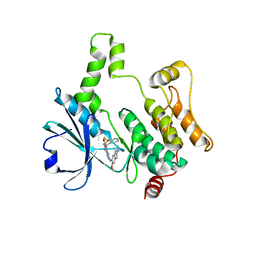 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 9f | | Descriptor: | 5-amino-2-((6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino)-8-(2-(trifluoromethyl)benzyl)pyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Huang, W.X, Liu, R, Ding, K. | | Deposit date: | 2023-12-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of 5-aminopyrido[2,3-d]pyrimidin-7(8H)-one derivatives as new hematopoietic progenitor kinase 1 (HPK1) inhibitors.
Eur.J.Med.Chem., 269, 2024
|
|
8YDC
 
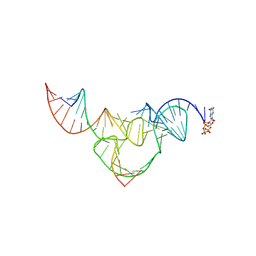 | | Crystal structure of a hammerhead ribozyme with pseudoknot | | Descriptor: | DNA/RNA (5'-R(*AP*CP*AP*UP*GP*UP*CP*U)-D(P*C)-R(P*UP*GP*GP*GP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, ribozyme strand | | Authors: | Liu, Y, Zhan, X. | | Deposit date: | 2024-02-20 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme.
Nat Commun, 15, 2024
|
|
7D4B
 
 | | Crystal structure of 4-1BB in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7CZD
 
 | | Crystal structure of PD-L1 in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Programmed cell death 1 ligand 1, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7DCB
 
 | |
7DCW
 
 | |
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKK
 
 | | De novo design protein XM2H | | Descriptor: | De novo design protein XM2H | | Authors: | Bin, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKO
 
 | | De novo design protein AM2M | | Descriptor: | de novo designed protein AM2M | | Authors: | Bin, H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
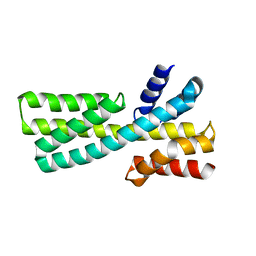 | |
7DGY
 
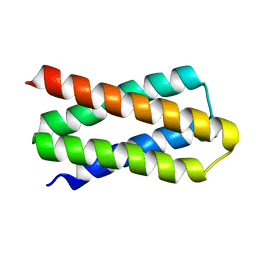 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DH1
 
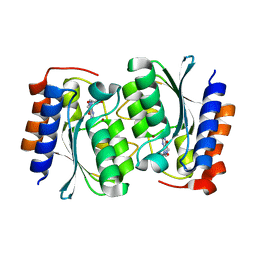 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM5
 
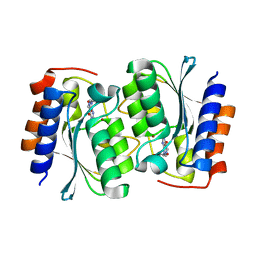 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with guanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DQN
 
 | |
7DM6
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase complexed with crotonoside | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1H-purin-2-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DGC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 2'-O-Methylguanosine | | Descriptor: | 9-[(2R,3R,4R)-5-(hydroxymethyl)-3-methoxy-4-oxidanyl-oxolan-2-yl]-3H-purine-2,6-dione, Guanosine deaminase, SODIUM ION, ... | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7EO0
 
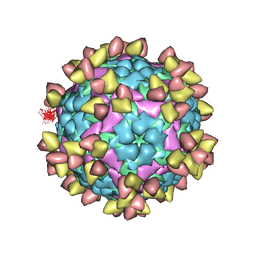 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7FBD
 
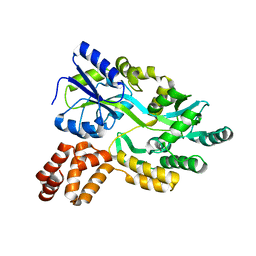 | | De novo design protein D53 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D53 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBB
 
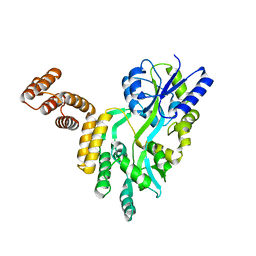 | | De novo design protein D12 with MBP tag | | Descriptor: | Maltodextrin-binding protein,de novo designed protein D12 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBC
 
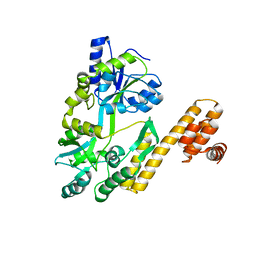 | | De novo design protein D22 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D22 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DBF
 
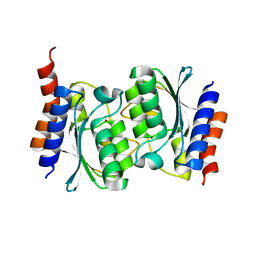 | |
7DC9
 
 | |
