4HC5
 
 | |
4HAM
 
 | | Crystal Structure of Transcriptional Antiterminator from Listeria monocytogenes EGD-e | | Descriptor: | GLYCEROL, Lmo2241 protein, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal Structure of Transcriptional Antiterminator from Listeria monocytogenes EGD-e
To be Published
|
|
4GUD
 
 | | Crystal Structure of Amidotransferase HisH from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Crystal Structure of Amidotransferase HisH from Vibrio cholerae.
To be Published
|
|
4GL0
 
 | | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lmo0810 protein, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-13 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes
To be Published
|
|
4GXT
 
 | | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548 | | Descriptor: | GLYCEROL, SULFATE ION, a conserved functionally unknown protein | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548
To be Published
|
|
4GS5
 
 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
3F1B
 
 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3F6O
 
 | | Crystal structure of ArsR family transcriptional regulator, RHA00566 | | Descriptor: | BROMIDE ION, Probable transcriptional regulator, ArsR family protein | | Authors: | Dong, A, Xu, X, Zheng, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ArsR family transcriptional regulator, RHA00566
To be Published
|
|
4FCA
 
 | | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. | | Descriptor: | Conserved domain protein, IMIDAZOLE, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames.
To be Published
|
|
4E2G
 
 | | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Cupin 2 conserved barrel domain protein, NICKEL (II) ION, ... | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-08 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus
To be Published
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
4E4Y
 
 | | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | GLYCEROL, SULFATE ION, Short chain dehydrogenase family protein | | Authors: | Zhang, R, Zhou, M, Tan, K, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4G6Q
 
 | | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Tan, K, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of a functionally unknown protein Kfla_6221 from Kribbella flavida DSM 17836, CASP Target
To be Published
|
|
4G6X
 
 | | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila. | | Descriptor: | GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila.
To be Published
|
|
4HG2
 
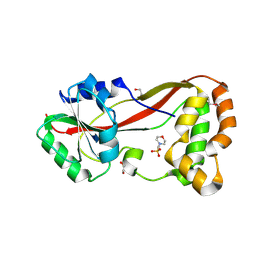 | | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Methyltransferase type 11 | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans.
TO BE PUBLISHED
|
|
3FBS
 
 | | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Oxidoreductase, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens
To be Published, 2008
|
|
4GQT
 
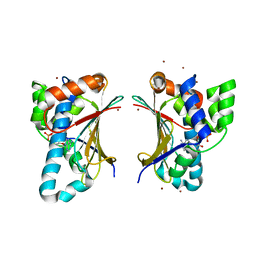 | | N-terminal domain of C. elegans Hsp90 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90, ZINC ION | | Authors: | Osipiuk, J, Chhor, G, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-terminal domain of C. elegans Hsp90
To be Published
|
|
4GYQ
 
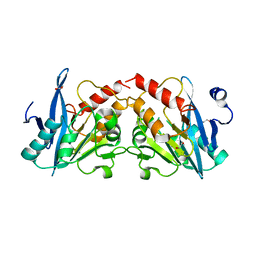 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4HAO
 
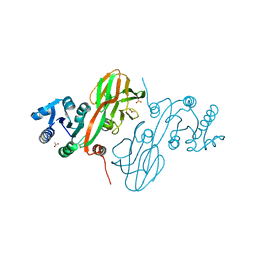 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92 | | Descriptor: | ACETIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92
To be Published
|
|
2APL
 
 | | Crystal structure of protein PG0816 from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG0816 | | Authors: | Chang, C, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of protein PG0816 from Porphyromonas gingivalis
To be Published
|
|
3F5R
 
 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
4HB7
 
 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
4GPN
 
 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4HCJ
 
 | | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kim, Y, Bigelow, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii
To be Published
|
|
2AG8
 
 | | NADP complex of Pyrroline-5-carboxylate reductase from Neisseria meningitidis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, pyrroline-5-carboxylate reductase | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes.
J.Mol.Biol., 354, 2005
|
|
