7DOK
 
 | |
8J5P
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 2,000lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8J5O
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 100lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
5ILA
 
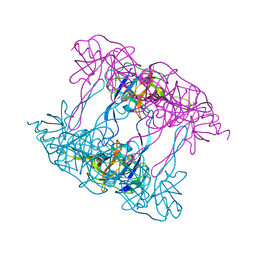 | | Deg9 protease domain | | Descriptor: | Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5ILB
 
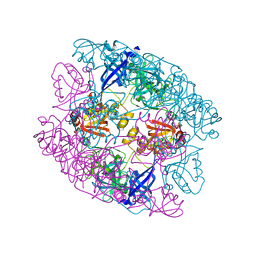 | | Crystal structure of protease domain of Deg2 linked with the PDZ domain of Deg9 | | Descriptor: | Protease Do-like 2, chloroplastic,Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
4E2A
 
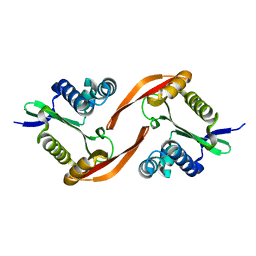 | |
5IL9
 
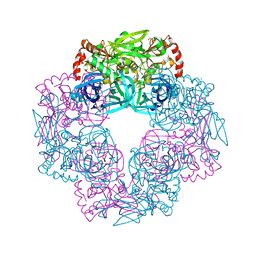 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
8H97
 
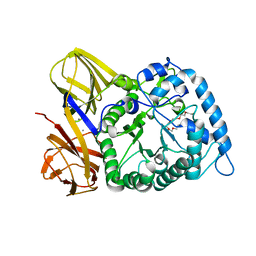 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
2PE2
 
 | |
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
4QR5
 
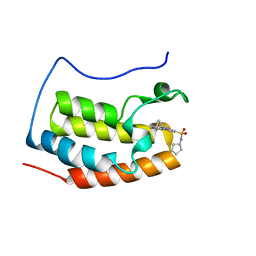 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR3
 
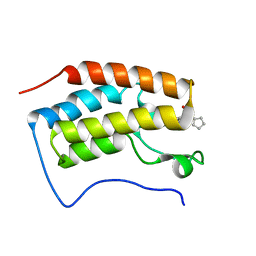 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
6ML6
 
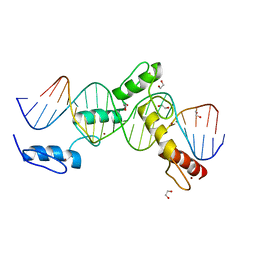 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML5
 
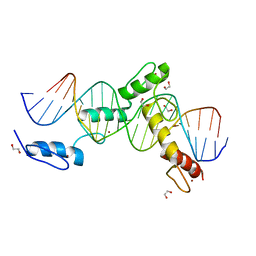 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML4
 
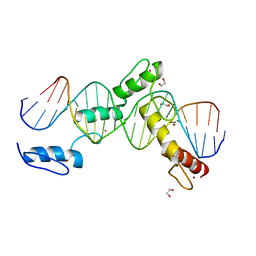 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML2
 
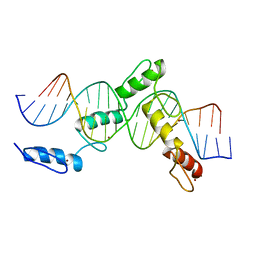 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
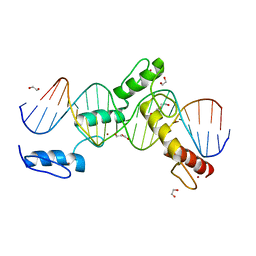 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML3
 
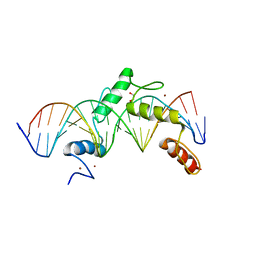 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
7E35
 
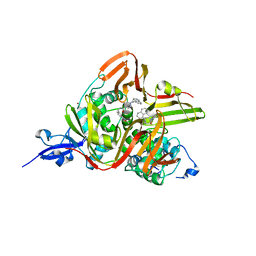 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
7D6H
 
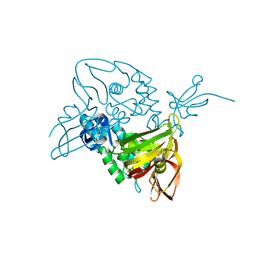 | |
7XGK
 
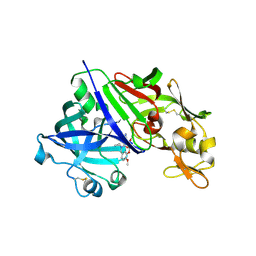 | | Human renin in complex with compound1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
