2O4M
 
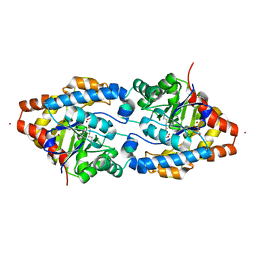 | | Structure of Phosphotriesterase mutant I106G/F132G/H257Y | | Descriptor: | ACETIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Phosphotriesterase mutant I106G/F132G/H257Y
To be Published
|
|
2OYP
 
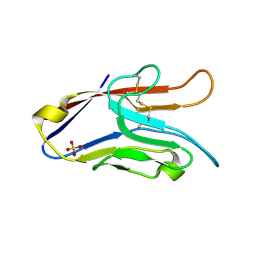 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
2Q1M
 
 | |
2IEC
 
 | | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri | | Descriptor: | MAGNESIUM ION, Uncharacterized protein conserved in archaea | | Authors: | Bonanno, J.B, Ramagopal, U.A, Dickey, M, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri
TO BE PUBLISHED
|
|
2IF7
 
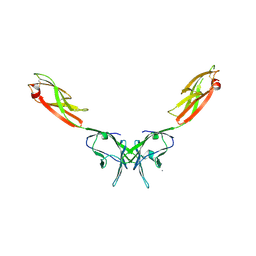 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
3GMF
 
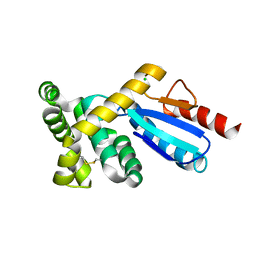 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3G7U
 
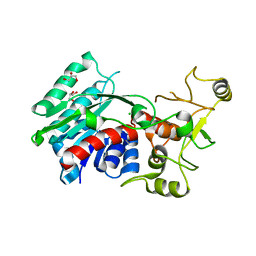 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GD5
 
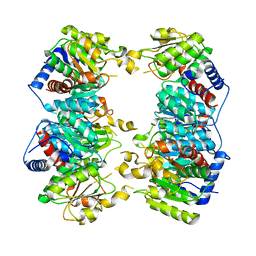 | | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Ramagopal, U.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus
To be Published
|
|
3GRZ
 
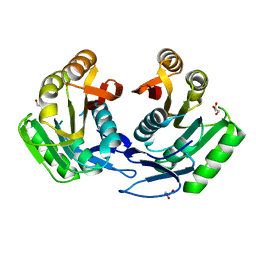 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3OCQ
 
 | |
3ODG
 
 | | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3EEY
 
 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
3CJP
 
 | | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum | | Descriptor: | Predicted amidohydrolase, dihydroorotase family, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U.A, Bonanno, J.B, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum.
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3E05
 
 | | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase FROM Geobacter metallireducens GS-15 | | Descriptor: | CHLORIDE ION, GLYCEROL, Precorrin-6Y C5,15-methyltransferase (Decarboxylating) | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Dickey, M, Hu, S, Maletic, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase from Geobacter metallireducens
To be Published
|
|
3FOM
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide IQQSIERL | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3FC0
 
 | | 1.8 A crystal structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells | | Descriptor: | ACETATE ION, GITR ligand | | Authors: | Chattopadhyay, K, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2008-11-20 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.8 A structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3E18
 
 | | CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-02 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua
To be Published
|
|
3FON
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | Descriptor: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
3FOL
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFERI | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, MHC | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
Int.Immunol., 22, 2010
|
|
3EBV
 
 | | Crystal structure of putative Chitinase A from Streptomyces coelicolor. | | Descriptor: | Chinitase A, SULFATE ION | | Authors: | Vigdorovich, V, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative Chitinase A from Streptomyces coelicolor
To be Published
|
|
3GHY
 
 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
