3STV
 
 | |
3STU
 
 | |
3STT
 
 | |
3STW
 
 | |
3W8W
 
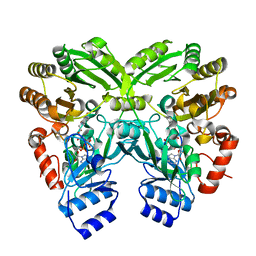 | | The crystal structure of EncM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W35
 
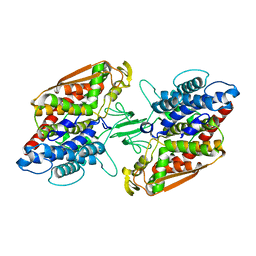 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W8Z
 
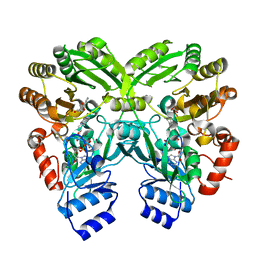 | | The complex structure of EncM with hydroxytetraketide | | Descriptor: | (7S)-7-hydroxy-1-phenyloctane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W8X
 
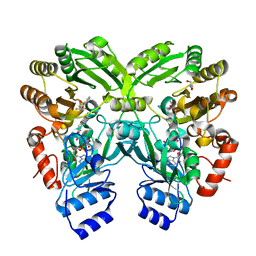 | | The complex structure of EncM with trifluorotriketide | | Descriptor: | 6,6,6-trifluoro-1-phenylhexane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W36
 
 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
2QW8
 
 | | Structure of Eugenol Synthase from Ocimum basilicum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Louie, G.V, Noel, J.P. | | Deposit date: | 2007-08-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase.
Plos One, 2, 2007
|
|
1M6E
 
 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
2R2G
 
 | | Structure of Eugenol Synthase from Ocimum basilicum complexed with EMDF | | Descriptor: | Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl (1S,2S)-2-(4-hydroxy-3-methoxyphenyl)cyclopropanecarboxylate | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2007-08-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase
Plos One, 2, 2007
|
|
2QX7
 
 | |
2QZZ
 
 | | Structure of Eugenol Synthase from Ocimum basilicum | | Descriptor: | Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl (1S,2S)-2-(4-hydroxy-3-methoxyphenyl)cyclopropanecarboxylate | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2007-08-17 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase
Plos One, 2, 2007
|
|
1KYZ
 
 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
1KYW
 
 | | Crystal Structure Analysis of Caffeic Acid/5-hydroxyferulic acid 3/5-O-methyltransferase in complex with 5-hydroxyconiferaldehyde | | Descriptor: | 5-(3,3-DIHYDROXYPROPENY)-3-METHOXY-BENZENE-1,2-DIOL, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
2Q6K
 
 | | SalL with adenosine | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, chlorinase | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
1ND7
 
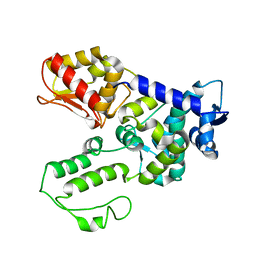 | | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase | | Descriptor: | WW domain-containing protein 1 | | Authors: | Verdecia, M.A, Joaziero, C.A.P, Wells, N.J, Ferrer, J.-L, Bowman, M.E, Hunter, T, Noel, J.P. | | Deposit date: | 2002-12-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Mol.Cell, 11, 2003
|
|
2Q6L
 
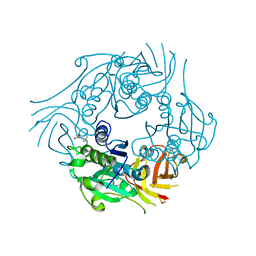 | | SalL double mutant Y70T/G131S with CLDA and L-MET | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Hypothetical protein, METHIONINE | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2R6J
 
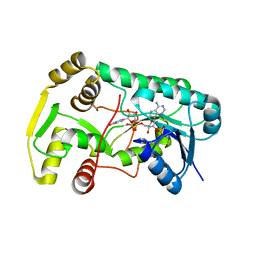 | |
1P15
 
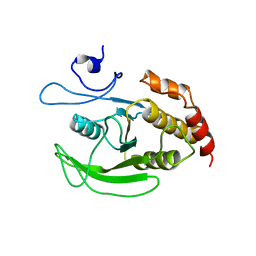 | |
2QYS
 
 | |
1OSH
 
 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
1QLV
 
 | |
1SUI
 
 | | Alfalfa caffeoyl coenzyme A 3-O-methyltransferase | | Descriptor: | CALCIUM ION, Caffeoyl-CoA O-methyltransferase, FERULOYL COENZYME A, ... | | Authors: | Ferrer, J.-L, Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Alfalfa Caffeoyl Coenzyme A 3-O-Methyltransferase
Plant Physiol., 137, 2005
|
|
