6BH7
 
 | | Phosphotriesterase variant R18+254S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18+254S
To Be Published
|
|
6BHK
 
 | | Phosphotriesterase variant R18deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase, ZINC ION | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18deltaL7
To Be Published
|
|
6BM9
 
 | |
4PBE
 
 | | Phosphotriesterase Variant Rev6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR6, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
5EH9
 
 | | Indirect contributions of mutations underlie optimization of new enzyme function | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLYCEROL, N-acyl homoserine lactonase AiiA, ... | | Authors: | Hong, N.-S, Jackson, C.J, Tokuriki, N, Yang, G, Baier, F. | | Deposit date: | 2015-10-28 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Conformational Tinkering Drives Evolution of a Promiscuous Activity through Indirect Mutational Effects.
Biochemistry, 55, 2016
|
|
6BQE
 
 | |
6BHL
 
 | | Phosphotriesterase variant S5deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant S5deltaL7
To Be Published
|
|
4PBF
 
 | | Phosphotriesterase variant Rev12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR12, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
5HMF
 
 | |
5HIF
 
 | | Crystal structure of a reconstructed lactonase ancestor, Anc1-MPH, of the bacterial methyl parathion hydrolase, MPH. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ZINC ION, ... | | Authors: | Baier, F, Carr, P.D, Jackson, C.J. | | Deposit date: | 2016-01-11 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | to be published
To Be Published
|
|
5HPQ
 
 | |
5HME
 
 | |
5HMD
 
 | |
7KW3
 
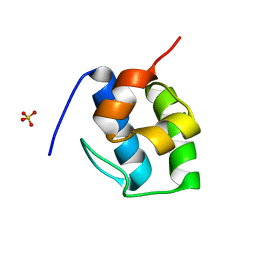 | | Non Ribosomal PCP domain | | Descriptor: | PCP domain, SULFATE ION | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KL8
 
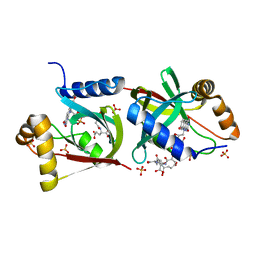 | | Structure of F420 binding protein Rv1558 from Mycobacterium tuberculosis with F420 bound | | Descriptor: | COENZYME F420, COENZYME F420-3, Deazaflavin-dependent nitroreductase, ... | | Authors: | Lee, B.M, Tan, L.L, Jackson, C.J. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Potency boost of a Mycobacterium tuberculosis dihydrofolate reductase inhibitor by multienzyme F 420 H 2 -dependent reduction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KW0
 
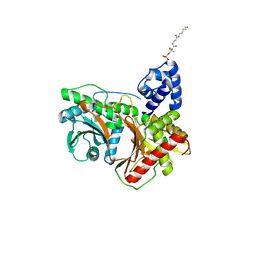 | | Non-ribosomal didomain (stabilised glycine-PCP-C) acceptor bound state | | Descriptor: | N-{2-[(2-aminoethyl)sulfanyl]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KVW
 
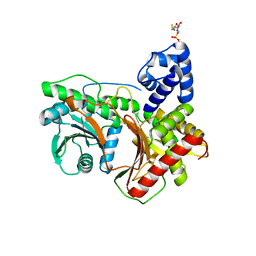 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KW2
 
 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state, R2577G | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
5BNC
 
 | | Structure of heme binding protein MSMEG_6519 from Mycobacterium smegmatis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ahmed, F.H, Carr, P.D, Jackson, C.J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
5C8V
 
 | |
7LNH
 
 | | S-adenosylmethionine synthetase co-crystallized with UppNHp | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-$l^{3}-sulfanyl]butanoic acid, 1,2-ETHANEDIOL, S-adenosylmethionine synthase isoform type-2 | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7LL3
 
 | | S-adenosylmethionine synthetase co-crystallized with UppNHp | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7LOW
 
 | | S-adenosylmethionine synthetase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7LOZ
 
 | | S-adenosylmethionine synthetase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-11 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7LNN
 
 | |
