2C2Z
 
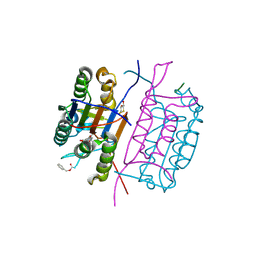 | | Crystal structure of caspase-8 in complex with aza-peptide Michael acceptor inhibitor | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL) -14-[4-(3,4-DIHYDROQUINOLIN-1(2H)-YL)-4-OXOBUTANOYL] -11-[(1R)-1-HYDROXYETHYL]-5-(2-METHYLPROPYL)-3,6,9,12-TETRAOXO -1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A.J, Mikolajczyk, J, Salvesen, G.S, Powers, J.C, Gruetter, M.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-09-20 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2CDR
 
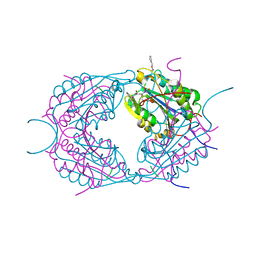 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EXPOXIDE, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Gruetter, M.G. | | Deposit date: | 2006-01-27 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors.
Biochemistry, 45, 2006
|
|
1FG2
 
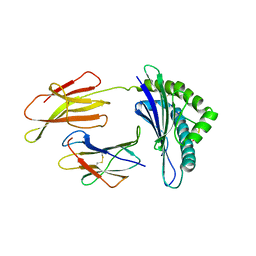 | | CRYSTAL STRUCTURE OF THE LCMV PEPTIDIC EPITOPE GP33 IN COMPLEX WITH THE MURINE CLASS I MHC MOLECULE H-2DB | | Descriptor: | BETA-2 MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Tissot, A.C, Ciatto, C, Mittl, P.R.E, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2000-07-27 | | Release date: | 2000-10-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.754 Å) | | Cite: | Viral escape at the molecular level explained by quantitative T-cell receptor/peptide/MHC interactions and the crystal structure of a peptide/MHC complex.
J.Mol.Biol., 302, 2000
|
|
3N2W
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA from Sphingosinicella xenopeptidilytica | | Descriptor: | Beta-peptidyl aminopeptidase, GLYCEROL, SULFATE ION | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P, Gruetter, M.G. | | Deposit date: | 2010-05-19 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Autoproteolytic and catalytic mechanisms for the beta-aminopeptidase BapA--a member of the Ntn hydrolase family.
Structure, 20, 2012
|
|
3N33
 
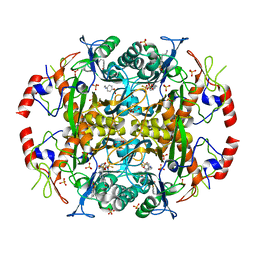 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with pefabloc SC (AEBSF) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P, Gruetter, M.G. | | Deposit date: | 2010-05-19 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoproteolytic and catalytic mechanisms for the beta-aminopeptidase BapA--a member of the Ntn hydrolase family.
Structure, 20, 2012
|
|
3N5I
 
 | | Crystal structure of the precursor (S250A mutant) of the N-terminal beta-aminopeptidase BapA | | Descriptor: | Beta-peptidyl aminopeptidase, GLYCEROL, SULFATE ION | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P, Gruetter, M.G. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoproteolytic and catalytic mechanisms for the beta-aminopeptidase BapA--a member of the Ntn hydrolase family.
Structure, 20, 2012
|
|
1PII
 
 | |
3GA4
 
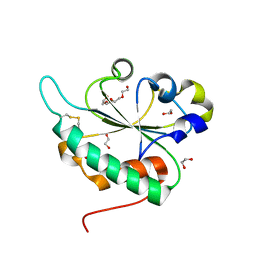 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G9B
 
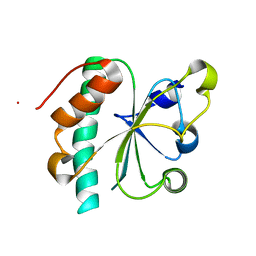 | | Crystal structure of reduced Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G7Y
 
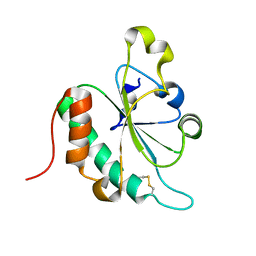 | | Crystal structure of oxidized Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HBX
 
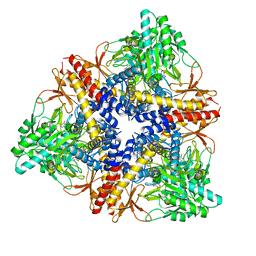 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
1N3N
 
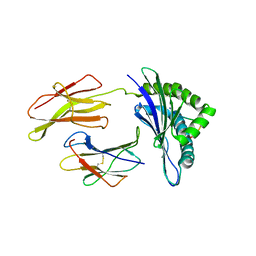 | | Crystal structure of a mycobacterial hsp60 epitope with the murine class I MHC molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Capitani, G, Tissot, A.C, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2002-10-29 | | Release date: | 2003-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of mycobacterial
and murine hsp60 epitopes in
complex with the class I MHC
molecule H-2D(b)
FEBS Lett., 543, 2003
|
|
1JFU
 
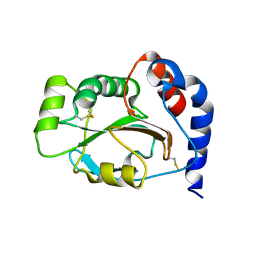 | | CRYSTAL STRUCTURE OF THE SOLUBLE DOMAIN OF TLPA FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN TLPA | | Authors: | Capitani, G, Rossmann, R, Sargent, D.F, Gruetter, M.G, Richmond, T.J, Hennecke, H. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties.
J.Mol.Biol., 311, 2001
|
|
1HIH
 
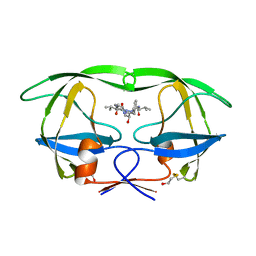 | |
1JJO
 
 | |
1KLX
 
 | |
1KTR
 
 | | Crystal Structure of the Anti-His Tag Antibody 3D5 Single-Chain Fragment (scFv) in Complex with a Oligohistidine peptide | | Descriptor: | Anti-his tag antibody 3d5 variable light chain, Peptide linker, Anti-his tag antibody 3d5 variable heavy chain, ... | | Authors: | Kaufmann, M, Lindner, P, Honegger, A, Blank, K, Tschopp, M, Capitani, G, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-01-17 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the anti-His tag antibody 3D5 single-chain fragment complexed to its antigen.
J.Mol.Biol., 318, 2002
|
|
1HII
 
 | |
2AXI
 
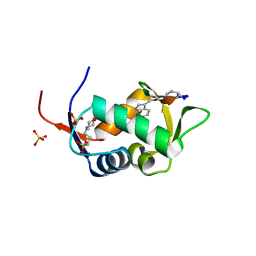 | | HDM2 in complex with a beta-hairpin | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, ... | | Authors: | Mittl, P.R.E, Fasan, R, Robinson, J, Gruetter, M.G. | | Deposit date: | 2005-09-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Studies in a Family of beta-Hairpin Protein Epitope Mimetic Inhibitors of the p53-HDM2 Protein-Protein Interaction.
Chembiochem, 7, 2006
|
|
1I1B
 
 | |
