6MJ9
 
 | |
4GNV
 
 | |
3EZN
 
 | |
3GP5
 
 | |
3GP3
 
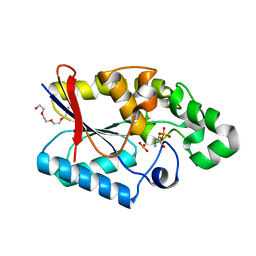 | |
3FDZ
 
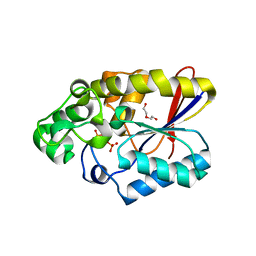 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3GW8
 
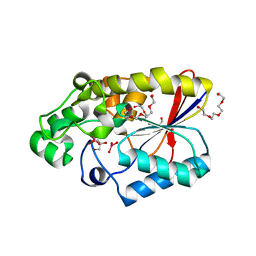 | |
5IF9
 
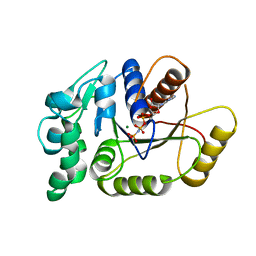 | |
5IHP
 
 | |
3LNT
 
 | |
6UH2
 
 | |
7UMP
 
 | | CRYSTAL STRUCTURE OF PHD2 CATALYTIC DOMAIN (CID 7465) IN COMPLEX WITH AKB-6548 AT 1.8 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Davie, D.R, Abendroth, J, Boyd, J. | | Deposit date: | 2022-04-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preclinical Characterization of Vadadustat (AKB-6548), an Oral Small Molecule Hypoxia-Inducible Factor Prolyl-4-Hydroxylase Inhibitor, for the Potential Treatment of Renal Anemia.
J.Pharmacol.Exp.Ther., 383, 2022
|
|
6CWA
 
 | |
6AUB
 
 | |
6AUA
 
 | |
4PFS
 
 | |
4MI2
 
 | |
1JLO
 
 | |
1JLP
 
 | |
6OP0
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (R)-{1-[(2,5-dimethylphenyl)methyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fairman, J.W, Davies, D.R, Foley, A, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
4W65
 
 | |
7MFC
 
 | |
1AS5
 
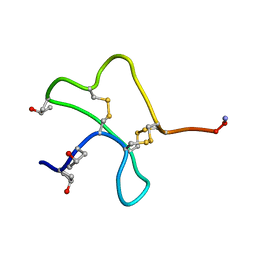 | | SOLUTION STRUCTURE OF CONOTOXIN Y-PIIIE FROM CONUS PURPURASCENS, NMR, 14 STRUCTURES | | Descriptor: | CONOTOXIN Y-PIIIE | | Authors: | Mitchell, S.S, Shon, K, Foster, M.P, Olivera, B.M, Ireland, C.M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of conotoxin psi-PIIIE, an acetylcholine gated ion channel antagonist.
Biochemistry, 37, 1998
|
|
6CFP
 
 | |
7SY9
 
 | |
