8Z27
 
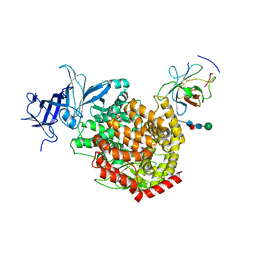 | | The structure of TGEV RBD and dog APN complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Sun, J.Q, Niu, S. | | Deposit date: | 2024-04-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species.
Plos Pathog., 21, 2025
|
|
8INP
 
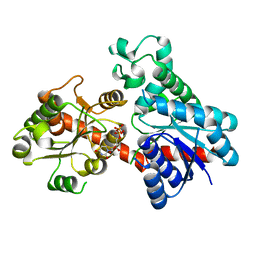 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT | | Descriptor: | Bc7OUGT, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8YZI
 
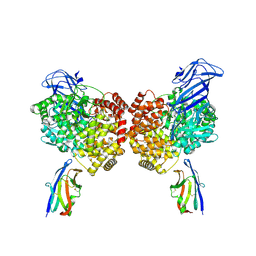 | | The structure of PDCoV RBD and dog APN complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Sun, J.Q, Niu, S. | | Deposit date: | 2024-04-07 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species.
Plos Pathog., 21, 2025
|
|
6ID3
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGPL ( A138S, V186G) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-08 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDD
 
 | | Crystal structure of H7 hemagglutinin mutant SH1-AVPL ( S138A, G186V, T221P, Q226L) from the influenza virus A/Shanghai/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
8YWG
 
 | |
8YWI
 
 | | The structure of ASFV DNA polymerase in replicating state | | Descriptor: | DNA polymerase, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kuai, L, Sun, J.Q, Peng, Q, Shi, Y. | | Deposit date: | 2024-03-31 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of DNA polymerase of African swine fever virus.
Nucleic Acids Res., 52, 2024
|
|
8YWM
 
 | | The structure of ASFV DNA polymerase in editing state | | Descriptor: | DNA polymerase, The primer strand, The template strand | | Authors: | Kuai, L, Sun, J.Q, Peng, Q, Shi, Y. | | Deposit date: | 2024-03-31 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of DNA polymerase of African swine fever virus.
Nucleic Acids Res., 52, 2024
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7D86
 
 | | Crystal Structure of zebrafishPHF14-PZP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
7D8A
 
 | | Crystal Structure of H3(1-13)/PHF14-PZP fusion protein | | Descriptor: | CALCIUM ION, Gene for histone H3 (germline gene), PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
7D87
 
 | | Crystal Structure of zebrafish PHF14-PZP in complex with H3(1-25) | | Descriptor: | CALCIUM ION, Gene for histone H3 (germline gene), PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
