4WEK
 
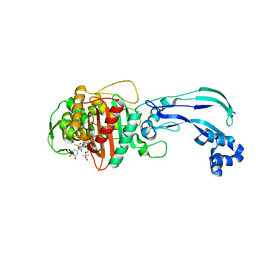 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WF1
 
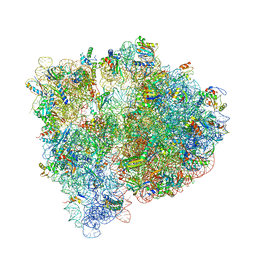 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WZ4
 
 | | Crystal structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, GLYCEROL, {(3R)-6-[(3-amino-1,2,4-thiadiazol-5-yl)oxy]-1-hydroxy-4,5-dimethyl-1,3-dihydro-2,1-benzoxaborol-3-yl}acetic acid | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
4X7D
 
 | |
1XDL
 
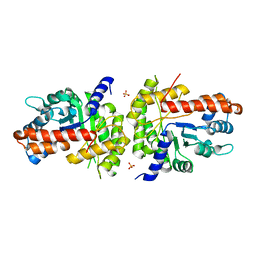 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 277K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
1XDM
 
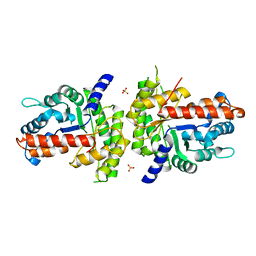 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 291K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
5TCZ
 
 | |
1ZBN
 
 | |
7T38
 
 | |
7T37
 
 | | Activated state of 2-APB and CBD-bound wildtype rat TRPV2 in nanodiscs | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2, cannabidiol | | Authors: | Pumroy, R.A, Protopopova, A.D, Gallo, P.N, Moiseenkova-Bell, V.Y. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into TRPV2 activation by small molecules.
Nat Commun, 13, 2022
|
|
4XMB
 
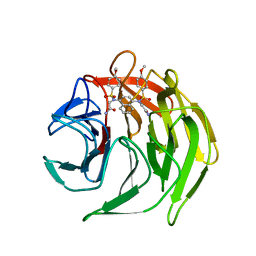 | | Crystal structure of 2,2'-(naphthalene-1,4-diylbis(((4-methoxyphenyl)sulfonyl)azanediyl))diacetamide bound to human Keap1 Kelch domain | | Descriptor: | 2,2'-(naphthalene-1,4-diylbis(((4-methoxyphenyl)sulfonyl)azanediyl))diacetamide, Kelch-like ECH-associated protein 1 | | Authors: | Luciano, J.P, Ryuzoji, A.F, Mesecar, A.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators.
Eur.J.Med.Chem., 103, 2015
|
|
6EWB
 
 | |
6CCX
 
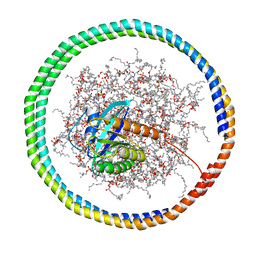 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6DHC
 
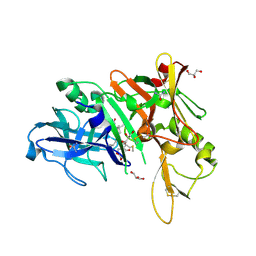 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | Descriptor: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Mesecar, A.D, Lendy, E.K. | | Deposit date: | 2018-05-19 | | Release date: | 2018-07-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8Q9Z
 
 | |
8QA2
 
 | |
8QA1
 
 | |
8QA3
 
 | |
8QA0
 
 | |
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6D
 
 | | 10 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6C
 
 | | 1 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
6W44
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
