2NRD
 
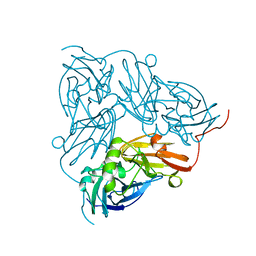 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
2O1C
 
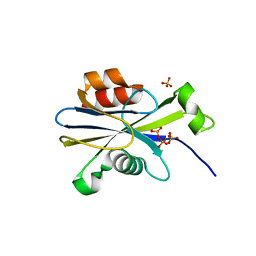 | |
2O5W
 
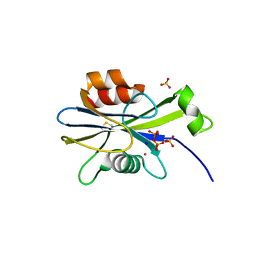 | | Structure of the E. coli dihydroneopterin triphosphate pyrophosphohydrolase in complex with Sm+3 and pyrophosphate | | Descriptor: | PYROPHOSPHATE, SAMARIUM (III) ION, SODIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2006-12-06 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis.
Structure, 15, 2007
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
2P6B
 
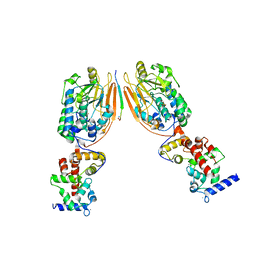 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
2PH0
 
 | | Crystal structure of the Q6D2T7_ERWCT protein from Erwinia carotovora. NESG target EwR41. | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Chen, C.-X, Cunningham, K, Ma, L.-C, Owens, L, Fang, Y, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-04-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Q6D2T7_ERWCT protein from Erwinia carotovora.
To be Published
|
|
2MHR
 
 | |
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1NIF
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIE
 
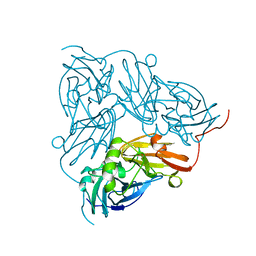 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1L9M
 
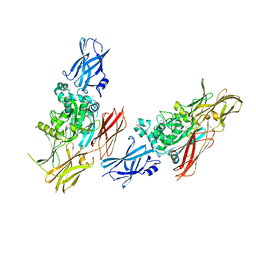 | |
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1NMC
 
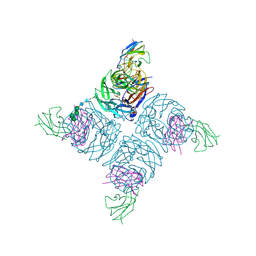 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 15 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
1L9N
 
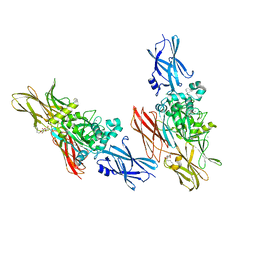 | |
1NIC
 
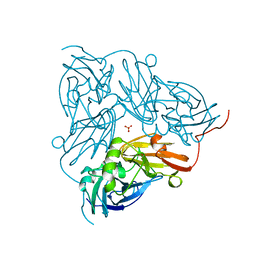 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, SULFATE ION | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NID
 
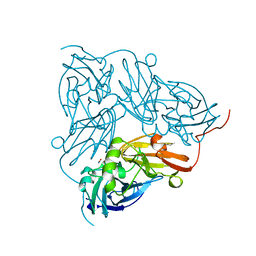 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIA
 
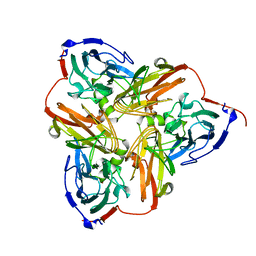 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1Q3A
 
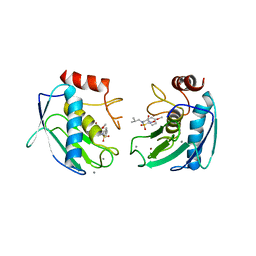 | | Crystal structure of the catalytic domain of human matrix metalloproteinase 10 | | Descriptor: | CALCIUM ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, Stromelysin-2, ... | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-07-29 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic domain of human matrix metalloproteinase 10.
J.Mol.Biol., 336, 2004
|
|
1DCD
 
 | | DESULFOREDOXIN COMPLEXED WITH CD2+ | | Descriptor: | CADMIUM ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Romao, M.J. | | Deposit date: | 1999-03-20 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1DE7
 
 | |
1EX0
 
 | | HUMAN FACTOR XIII, MUTANT W279F ZYMOGEN | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII A CHAIN, PHOSPHATE ION, ... | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-28 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To be Published
|
|
1FKZ
 
 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
1BT7
 
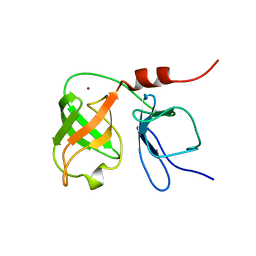 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
1FKY
 
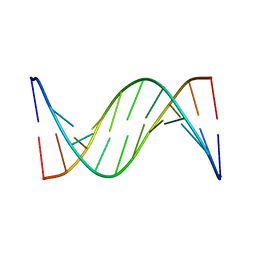 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
