8XE5
 
 | | norbelladine 4'-O-methyltransferase complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Norbelladine 4'-O-methyltransferase, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XE3
 
 | |
8XE0
 
 | | O-methyltransferase from Lycoris longituba M52W variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | MAGNESIUM ION, Protocatechuic aldehyde, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDV
 
 | |
8XE2
 
 | | O-methyltransferase from Lycoris longituba D230A variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protocatechuic aldehyde, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
6AJV
 
 | | Crystal structure of BRD4 in complex with isoliquiritigenin and DMSO (Cocktail No. 3) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
8Z8S
 
 | |
8ZFW
 
 | |
8Z8R
 
 | |
8Z8P
 
 | |
8ZYD
 
 | |
8ZYC
 
 | |
8WTR
 
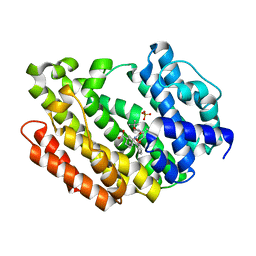 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH (1S,3R)-3-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6,8-dioxo-5,6,7,8,10,11-hexahydro-13H-12-oxa-5,9-diaza-benzocycloundecen-9-yl]-cyclohexanecarboxylic acid | | Descriptor: | (1~{S},3~{R})-3-[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3,5-bis(oxidanylidene)-9-oxa-2,6-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-6-yl]cyclohexane-1-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6AJY
 
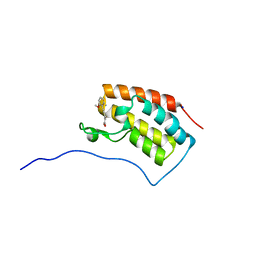 | | Crystal structure of BRD4 in complex with 2',4'-dihydroxy-2-methoxychalcone | | Descriptor: | 2',4'-dihydroxy-2-methoxychalcone, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
8Z8O
 
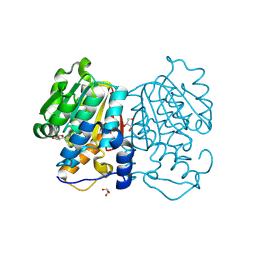 | |
8WTQ
 
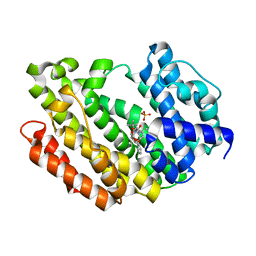 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH {1-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6-oxo-6,7,10,11-tetrahydro-5H,9H,13H-12-oxa-5,8-diaza-benzocycloundecene-8-carbonyl]-piperidin-4-yl}-acetic acid | | Descriptor: | 2-[1-[[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3-oxidanylidene-9-oxa-2,5-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-5-yl]carbonyl]piperidin-4-yl]ethanoic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6AJX
 
 | | Crystal structure of BRD4 in complex with isoliquiritigenin in the absence of DMSO | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJZ
 
 | | Joint nentron and X-ray structure of BRD4 in complex with colchicin | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, SODIUM ION | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.301 Å), X-RAY DIFFRACTION | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJW
 
 | | Crystal structure of BRD4 in complex with DMSO (Cocktail No. 4) | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6L2F
 
 | | Crystal structure of a cupin protein (tm1459, H54AH58A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, COPPER (II) ION, ... | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | Descriptor: | COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LOS
 
 | | Crystal structure of mouse PEDF in complex with heterotrimeric collagen model peptide. | | Descriptor: | Collagen model peptide, type I, alpha 1, ... | | Authors: | Kawahara, K, Maruno, T, Oki, H, Yoshida, T, Ohkubo, T, Koide, T, Kobayashi, Y. | | Deposit date: | 2020-01-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Spatiotemporal regulation of PEDF signaling by type I collagen remodeling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2RO0
 
 | | Solution structure of the knotted tudor domain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
