8WK7
 
 | |
5OBX
 
 | | Mycoplasma genitalium DnaK-NBD | | Descriptor: | Chaperone protein DnaK, GLYCEROL, SULFATE ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
3V82
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
5O4H
 
 | | HcgC from Methanococcus maripaludis cocrystallized with SAM and pyridinol | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, HcgC, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1PNN
 
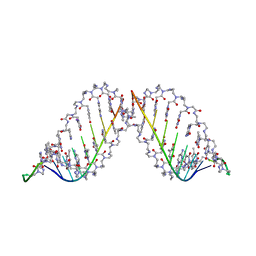 | | PEPTIDE NUCLEIC ACID (PNA) COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(GP*AP*AP*GP*AP*AP*GP*AP*G)-3'), PNA (NH2-P(*C*T*C*T*T*C*T*T*C-HIS-GLY-SER-SER-GLY-HIS-C*T*T*C*T*T*C*T*C)-COOH) | | Authors: | Betts, L, Veal, J.M. | | Deposit date: | 1995-10-13 | | Release date: | 1996-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Nucleic Acid Triple Helix Formed by a Peptide Nucleic Acid-DNA Complex
Science, 270, 1995
|
|
4BTU
 
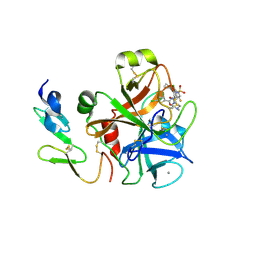 | | Factor Xa in complex with the dual thrombin-FXa inhibitor 57. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-chloro-5-fluoro-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-(4-methyl-piperazin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
3VB9
 
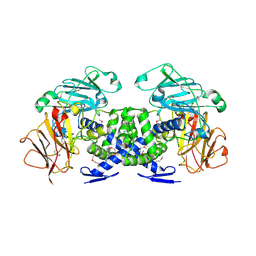 | | Crystal structure of VPA0735 from Vibrio parahaemolyticus in monoclinic form, NorthEast Structural Genomics target VpR109 | | Descriptor: | MAGNESIUM ION, Uncharacterized protein VPA0735 | | Authors: | Seetharaman, J, Neely, H, Cunningham, K, Ciccosanti, C, Bjelic, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of VPA0735 from Vibrio parahaemolyticus in monoclinic form, NorthEast Structural Genomics target VpR109
To be Published
|
|
8WKA
 
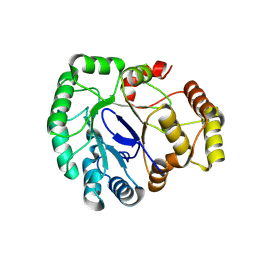 | |
3VCI
 
 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
3VCK
 
 | | Thaumatin by LB Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
1PWA
 
 | | Crystal structure of Fibroblast Growth Factor 19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fibroblast growth factor-19, GLYCEROL, ... | | Authors: | Harmer, N.J, Pellegrini, L, Chirgadze, D, Fernandez-Recio, J, Blundell, T.L. | | Deposit date: | 2003-07-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity.
Biochemistry, 43, 2004
|
|
4BPW
 
 | | Crystal structure of human primase bound to UTP | | Descriptor: | DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, MAGNESIUM ION, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BUU
 
 | | Crystal structure of human tankyrase 2 in complex with (4-(4-oxo-3,4- dihydroquinazolin-2-yl)phenyl)methanesulfonamide | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
4BTX
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-isopropylamino-2-phenyl-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, CALCIUM ION, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
1E6K
 
 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
1E7O
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25V, V44I, V58L MUTATIONS | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-31 | | Release date: | 2003-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Thermodynamic and Kinetic Analysis of the Folding Pathway of an SH3 Domain Entropically Stabilised by a Redesigned Hydrophobic Core
J.Mol.Biol., 328, 2003
|
|
4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4BS4
 
 | | Crystal structure of human tankyrase 2 in complex with 4'-isopropylflavone | | Descriptor: | 4'-ISOPROPYLFLAVONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Tankyrase Inhibiting Flavones with Increased Potency and Isoenzyme Selectivity.
J.Med.Chem., 56, 2013
|
|
4BTJ
 
 | | TTBK1 in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Xue, Y, Wan, P, Hillertz, P, Schweikart, F, Zhao, Y, Wissler, L, Dekker, N. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | X-Ray Structural Analysis of Tau-Tubulin Kinase 1 and its Interactions with Small Molecular Inhibitors.
Chemmedchem, 8, 2013
|
|
5O69
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
4BW1
 
 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-tert-butylphenyl)amino]-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-3-carboxylic acid, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
1FW4
 
 | |
