6T90
 
 | | OCT4-SOX2-bound nucleosome - SHL-6 | | Descriptor: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6RCQ
 
 | | PfRH5-binding monoclonal antibody R5.011 | | Descriptor: | ACETATE ION, CACODYLATE ION, R5.011 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCS
 
 | | PfRH5-binding monoclonal antibody R5.016 | | Descriptor: | ACETATE ION, R5.016 heavy chain, R5.016 light chain, ... | | Authors: | Alanine, D.W.G, Jamwal, A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
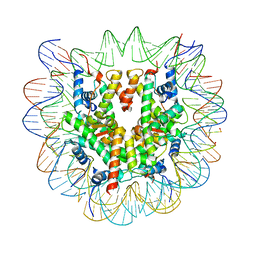 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | Descriptor: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
5AN7
 
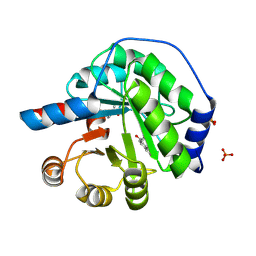 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | Authors: | Obexer, R, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2015-09-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
5DMM
 
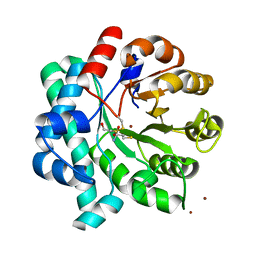 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Metallated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, BETA-MERCAPTOETHANOL, Homocysteine S-methyltransferase, ... | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
6RF8
 
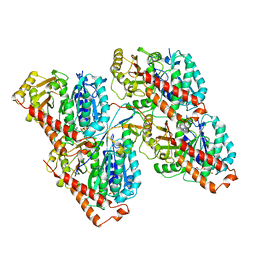 | |
5A5P
 
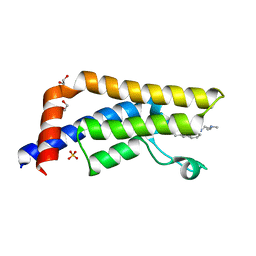 | | Crystal structure of human ATAD2 bromodomain in complex with 8-2-(dimethylamino)ethylamino-3-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(dimethylamino)ethyl]amino}-3-methyl-1,2-dihydroquinolin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
6TN6
 
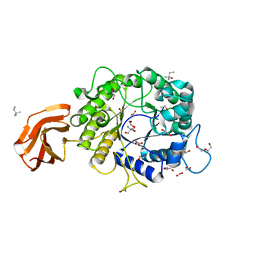 | | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | da Silva, V.M, Squina, F.M, Sperenca, M, Martin, L, Muniz, J.R.C, Garcia, W, Nicolet, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structure of a modular hyperthermostable endo-beta-1,4-mannanase from Thermotoga petrophila: The ancillary immunoglobulin-like module is a thermostabilizing domain.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
5EBT
 
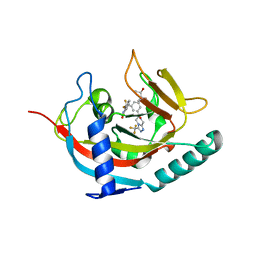 | | Tankyrase 1 with Phthalazinone 2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-[bis(fluoranyl)-[3-[[(6~{S})-6-methyl-3-(trifluoromethyl)-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazin-7-yl]carbonyl]phenyl]methyl]-2~{H}-phthalazin-1-one, Tankyrase-1, ... | | Authors: | Kazmirski, S.L, Johannes, J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5DML
 
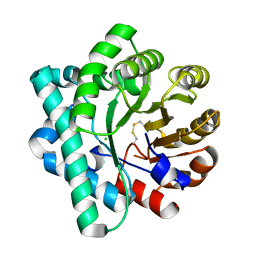 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Oxidized form | | Descriptor: | CHLORIDE ION, Homocysteine S-methyltransferase | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
5DMN
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Apo form | | Descriptor: | Homocysteine S-methyltransferase, SULFATE ION | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
7SH6
 
 | |
7T49
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4B
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T40
 
 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T42
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4A
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 11c | | Descriptor: | (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
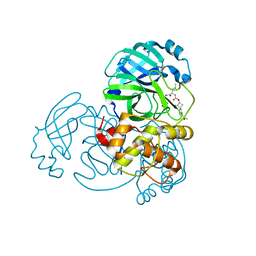 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T45
 
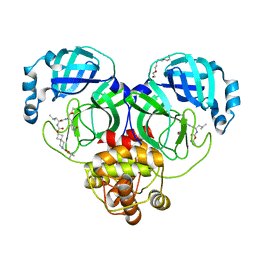 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 7c | | Descriptor: | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Kankanamalage, A.C.G, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
