6M07
 
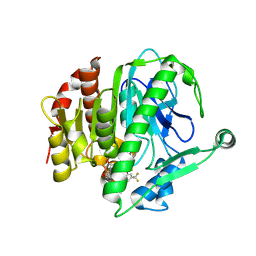 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
6M39
 
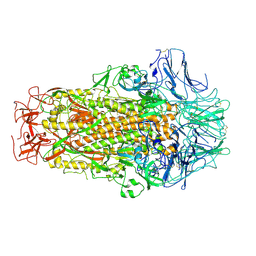 | | Cryo-EM structure of SADS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.
J.Virol., 94, 2020
|
|
8JJB
 
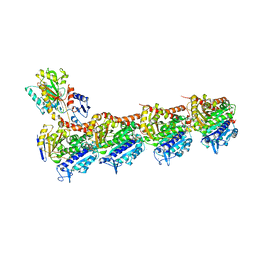 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
6M06
 
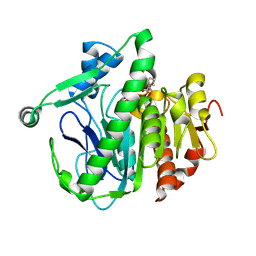 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(Z)-1,3-bis(oxidanyl)-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6UT9
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 | | Descriptor: | Outer capsid protein VP4 | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5D0N
 
 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
4G6H
 
 | | Crystal structure of NDH with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
5D1F
 
 | | Crystal structure of maize PDRP bound with AMP and Hg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-04 | | Release date: | 2016-02-24 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
5Z08
 
 | |
4G6G
 
 | | Crystal structure of NDH with TRT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, MAGNESIUM ION, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
5Z1W
 
 | |
1ZVS
 
 | | Crystal structure of the first class MHC mamu and Tat-Tl8 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Tat-Tl8 | | Authors: | Lou, Z, Chu, F, Gao, G.F, Rao, Z. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-13 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape.
J Immunol., 178, 2007
|
|
5Z07
 
 | | Crystal structure of centromere protein Cenp-I | | Descriptor: | Cenp-I | | Authors: | Tian, W, Hu, L.Q, He, X. | | Deposit date: | 2017-12-18 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment.
Nucleic Acids Res., 47, 2019
|
|
6V6R
 
 | | Crystal Structure of a Bromine Derivatized Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry. | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4G74
 
 | | Crystal structure of NDH with Quinone | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4K8V
 
 | | Structure of cyclic GMP-AMP Synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K1I
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
7TEU
 
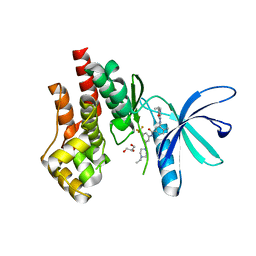 | | Crystal structure of JAK2 JH1 with type II inhibitor YLIU-04-105-1 | | Descriptor: | 3-{(4S)-2-[(cyclopropanecarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Hubbard, S.R. | | Deposit date: | 2022-01-05 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New scaffolds for type II JAK2 inhibitors overcome the acquired G993A resistance mutation.
Cell Chem Biol, 30, 2023
|
|
6W1K
 
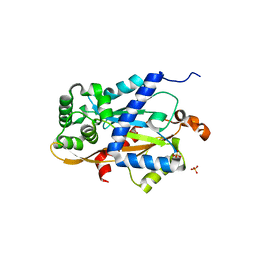 | | Crystal structure of the hydroxyglutarate synthase in complex with 2-oxoadipate from Oryza sativa | | Descriptor: | 2-OXOADIPIC ACID, Hydroxyglutarate synthase, NICKEL (II) ION, ... | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
6U95
 
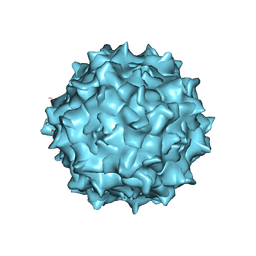 | | Adeno-associated virus strain AAVhu.37 capsid icosahedral structure | | Descriptor: | Capsid protein VP1 | | Authors: | Kaelber, J.T, Yost, S.A, Firlar, E, Mercer, A.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structure of the AAVhu.37 capsid by cryoelectron microscopy.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4G73
 
 | | Crystal structure of NDH with NADH and Quinone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
8WP1
 
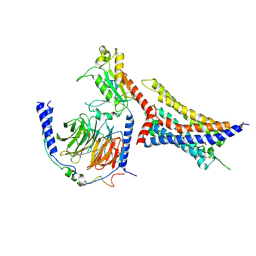 | | Cryo-EM structure of SUCR1 in complex with cis-epoxysuccinic acid and Gi proteins | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, A, Ye, R.D. | | Deposit date: | 2023-10-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into ligand recognition and activation of the succinate receptor SUCNR1.
Cell Rep, 43, 2024
|
|
6W1G
 
 | | Crystal structure of the hydroxyglutarate synthase from Pseudomonas putida | | Descriptor: | Hydroxyglutarate synthase, NICKEL (II) ION | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
8WOG
 
 | | Cryo-EM structure of SUCR1 in complex with succinate and Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Liu, A, Ye, R.D. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into ligand recognition and activation of the succinate receptor SUCNR1.
Cell Rep, 43, 2024
|
|
