2QSV
 
 | | Crystal structure of protein of unknown function from Porphyromonas gingivalis W83 | | Descriptor: | SODIUM ION, SULFATE ION, Uncharacterized protein | | Authors: | Binkowski, T.A, Duggan, E, Shui, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein of unknown function from Porphyromonas gingivalis W83.
TO BE PUBLISHED
|
|
2QRU
 
 | | Crystal structure of an alpha/beta hydrolase superfamily protein from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Cuff, M.E, Volkart, L, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-29 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of an alpha/beta hydrolase superfamily protein from Enterococcus faecalis.
TO BE PUBLISHED
|
|
2QX2
 
 | | Structure of the C-terminal domain of sex pheromone staph-cAM373 precursor from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Sex pheromone staph-cAM373 | | Authors: | Cuff, M.E, Mussar, K, Hatzos, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of sex pheromone staph-cAM373 precursor from Staphylococcus aureus.
TO BE PUBLISHED
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
2R41
 
 | |
2R2C
 
 | |
2QH9
 
 | |
2QHK
 
 | |
2QM3
 
 | | Crystal structure of a predicted methyltransferase from Pyrococcus furiosus | | Descriptor: | ACETIC ACID, CALCIUM ION, Predicted methyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a predicted methyltransferase from Pyrococcus furiosus.
TO BE PUBLISHED
|
|
2QMX
 
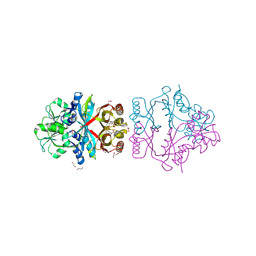 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2QSW
 
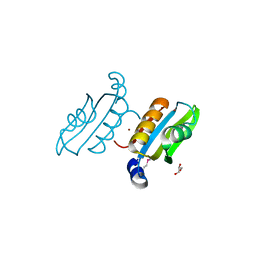 | | Crystal structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis | | Descriptor: | GLYCEROL, Methionine import ATP-binding protein metN 2, ZINC ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis.
To be Published
|
|
2QZI
 
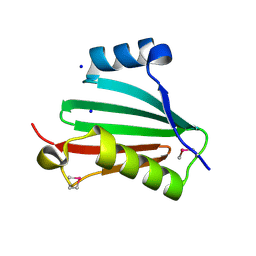 | | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311. | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311.
To be Published
|
|
2QSX
 
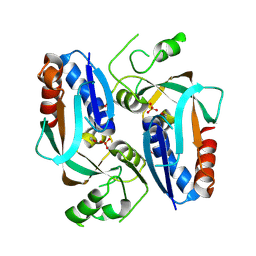 | | Crystal structure of putative transcriptional regulator LysR From Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator, LysR family, SULFATE ION | | Authors: | Wu, R, Abdullah, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Putative Transcriptional Regulator LysR From Vibrio parahaemolyticus.
To be Published
|
|
2R6H
 
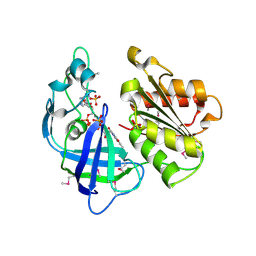 | | Crystal structure of the domain comprising the NAD binding and the FAD binding regions of the NADH:ubiquinone oxidoreductase, Na translocating, F subunit from Porphyromonas gingivalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH:ubiquinone oxidoreductase, Na translocating, ... | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Domain Comprising the Regions Binding NAD and FAD from the NADH:Ubiquinone Oxidoreductase, Na Translocating, F Subunit from Porphyromonas gingivalis.
To be Published
|
|
2NS0
 
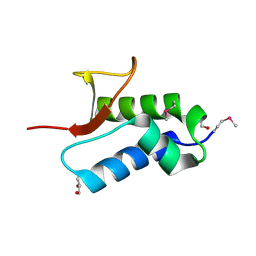 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
2RIR
 
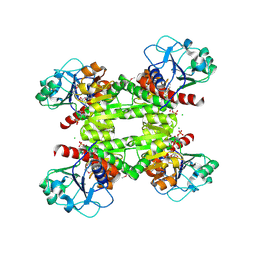 | | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis | | Descriptor: | CHLORIDE ION, Dipicolinate synthase, A chain, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis.
To be Published
|
|
2OB5
 
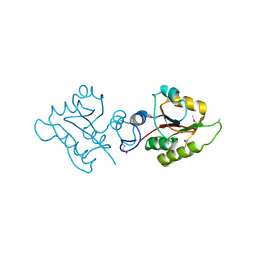 | | Crystal structure of protein Atu2016, putative sugar binding protein | | Descriptor: | Hypothetical protein Atu2016 | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of protein Atu2016, putative sugar binding protein
To be Published
|
|
2OI8
 
 | | Crystal structure of putative regulatory protein SCO4313 | | Descriptor: | Putative regulatory protein SCO4313 | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tetR family protein SCO4313
To be Published
|
|
2O8I
 
 | | Crystal structure of protein Atu2327 from Agrobacterium tumefaciens str. C58 | | Descriptor: | Hypothetical protein Atu2327 | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of protein Atu2327 from Agrobacterium tumefaciens str. C58
To be Published
|
|
4E4Y
 
 | | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | GLYCEROL, SULFATE ION, Short chain dehydrogenase family protein | | Authors: | Zhang, R, Zhou, M, Tan, K, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
5CNP
 
 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4EEI
 
 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with AMP and Succinate
To be Published
|
|
4EMY
 
 | |
4EAE
 
 | | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e | | Descriptor: | D-MALATE, Lmo1068 protein, SODIUM ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e
To be Published
|
|
4EAQ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate
To be Published
|
|
