6NPO
 
 | | Crystal structure of oligopeptide ABC transporter from Bacillus anthracis str. Ames (substrate-binding domain) | | Descriptor: | Oligopeptide ABC transporter, oligopeptide-binding protein, Unknown peptide ligand, ... | | Authors: | Michalska, K, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oligopeptide ABC transporter from
Bacillus anthracis str. Ames (substrate-binding domain)
To Be Published
|
|
6N7M
 
 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | Descriptor: | Hypothetical Protein CD630_05490 | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
6NFD
 
 | | beta-lactamase SHV-11 from Klebsiella pneumoniae strain NTUH-K2044 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, beta-lactamase SHV-11 | | Authors: | Osipiuk, J, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-19 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | beta-lactamase SHV-11 from Klebsiella pneumoniae strain NTUH-K2044
to be published
|
|
6NXI
 
 | | Flavin Transferase ApbE from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NHS
 
 | | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc
To Be Published
|
|
6NI0
 
 | | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Kim, Y, Wu, R, Endres, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis
To Be Published
|
|
6NJK
 
 | |
6NAU
 
 | | 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
4PF1
 
 | | Crystal structure of aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon | | Descriptor: | GLYCEROL, Peptidase S15/CocE/NonD, TRIETHYLENE GLYCOL | | Authors: | Michalska, K, Chhor, G, Fayman, K, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New aminopeptidase from "microbial dark matter" archaeon.
FASEB J., 29, 2015
|
|
2RK5
 
 | |
2PPW
 
 | |
2PMA
 
 | | Structural Genomics, the crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | ACETATE ION, FORMIC ACID, Uncharacterized protein | | Authors: | Tan, K, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
2VB1
 
 | | HEWL at 0.65 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LYSOZYME C, ... | | Authors: | Wang, J, Dauter, M, Alkire, R, Joachimiak, A, Dauter, Z. | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (0.65 Å) | | Cite: | Triclinic Lysozyme at 0.65 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6NJ1
 
 | |
6NHU
 
 | | Crystal Structure of the Beta Lactamase Class D YbxI from Agrobacterium fabrum | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Kim, Y, Welk, L, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbxI from Agrobacterium fabrum
To Be Published
|
|
6NI1
 
 | | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis
To Be Published
|
|
6NIQ
 
 | | Crystal Structure of the Putative Class A Beta-Lactamase PenP from Rhodopseudomonas palustris | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Crystal Structure of the Putative Class A Beta-Lactamase PenP from Rhodopseudomonas palustris
To Be Published
|
|
2QSA
 
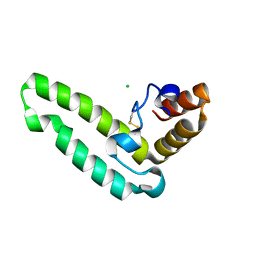 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
2QYB
 
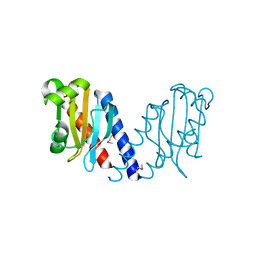 | |
2QRR
 
 | | Crystal structure of the soluble domain of the ABC transporter, ATP-binding protein from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, Methionine import ATP-binding protein metN | | Authors: | Kim, Y, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-28 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Soluble Domain of the ABC Transporter, ATP-binding Protein from Vibrio parahaemolyticus.
To be Published
|
|
2NNN
 
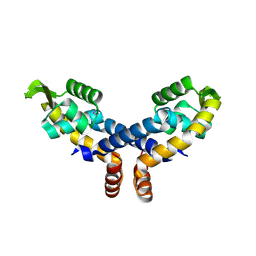 | | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Altamentova, S, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
2Q6J
 
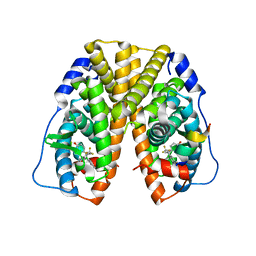 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
2QM2
 
 | | Putative HopJ type III effector protein from Vibrio parahaemolyticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, POTASSIUM ION, ... | | Authors: | Kim, Y, Chang, C, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Putative HopJ type III Effector Protein from Vibrio parahaemolyticus.
To be Published
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
