2RVM
 
 | |
2RVN
 
 | |
2RVL
 
 | |
3UNC
 
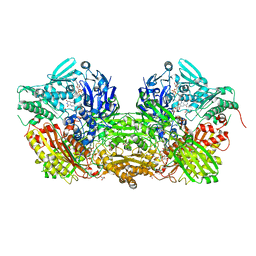 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNI
 
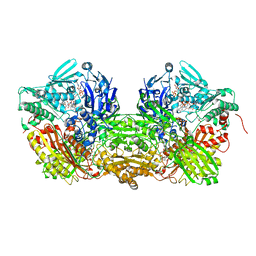 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NADH Bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNA
 
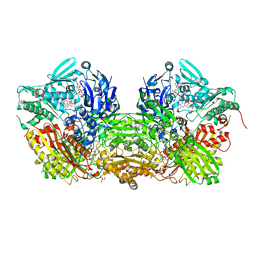 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NAD Bound | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3W0C
 
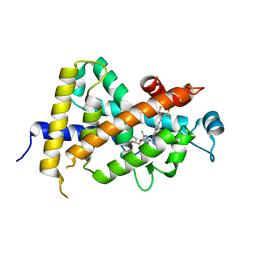 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[2-methyl-4-(3-{3-methyl-4-[(1E)-4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-en-1-yl]phenyl}pentan-3-yl)phenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | structure analysis of vitamin D receptor
To be Published
|
|
3W0A
 
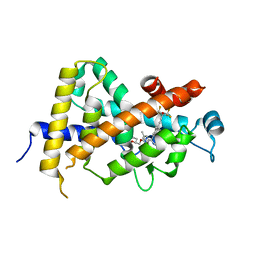 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[2-methyl-4-(3-{3-methyl-4-[4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-yn-1-yl]phenyl}pentan-3-yl)phenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | structure analysis of vitamin D3 receptor
To be Published
|
|
6JRQ
 
 | | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate synthetase, INOSINIC ACID | | Authors: | Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus HB8
To Be Published
|
|
3AZ1
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, {4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ2
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | 5-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ3
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[4-(3-{4-[(3S)-3-hydroxy-4,4-dimethylpentyl]-3-methylphenyl}pentan-3-yl)-2-methylphenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3W0Y
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, [3-fluoro-2'-methyl-4'-(3-{3-methyl-4-[(1E)-4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-en-1-yl]phenyl}pentan-3-yl)biphenyl-4-yl]acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure Analysis of Vitamin D receptor
TO BE PUBLISHED
|
|
5UND
 
 | |
2O3Z
 
 | | X-ray crystal structure of LpxC complexed with 3-heptyloxybenzoate | | Descriptor: | 3-(heptyloxy)benzoic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Christianson, D.W. | | Deposit date: | 2006-12-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Amphipathic benzoic acid derivatives: synthesis and binding in the hydrophobic tunnel of the zinc deacetylase LpxC.
Bioorg.Med.Chem., 15, 2007
|
|
2Z5E
 
 | | Crystal Structure of Proteasome Assembling Chaperone 3 | | Descriptor: | Proteasome Assembling Chaperone 3 | | Authors: | Okamoto, K, Kurimoto, E, Sakata, E, Suzuki, A, Yamane, T, Hirano, Y, Murata, S, Tanaka, K, Kato, K. | | Deposit date: | 2007-07-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3A2B
 
 | |
2Z2T
 
 | |
1US5
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2E7A
 
 | |
3A4P
 
 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2D6F
 
 | |
2DKF
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8, a RNA Degradation Protein of the Metallo-beta-lactamase Superfamily | | Descriptor: | ZINC ION, metallo-beta-lactamase superfamily protein | | Authors: | Ishikawa, I, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-10 | | Release date: | 2006-12-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-beta-lactamase superfamily
J.Biochem.(Tokyo), 140, 2006
|
|
6O1S
 
 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
6UEO
 
 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
