4YIT
 
 | |
4YHX
 
 | |
4Z1Z
 
 | |
4Z20
 
 | |
4YXX
 
 | |
4YY5
 
 | |
4YXZ
 
 | |
4YXY
 
 | |
1P6O
 
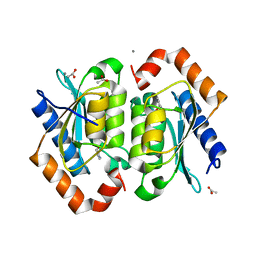 | | The crystal structure of yeast cytosine deaminase bound to 4(R)-hydroxyl-3,4-dihydropyrimidine at 1.14 angstroms. | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ireton, G.C, Black, M.E, Stoddard, B.L. | | Deposit date: | 2003-04-29 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.14 a crystal structure of yeast Cytosine deaminase. Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
Structure, 11, 2003
|
|
7SQ4
 
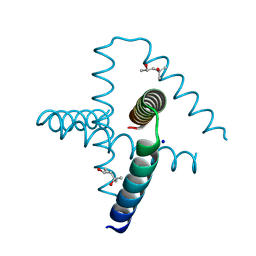 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ3
 
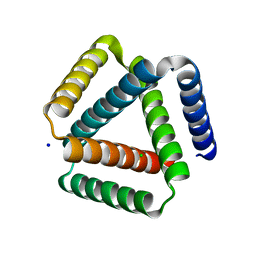 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ5
 
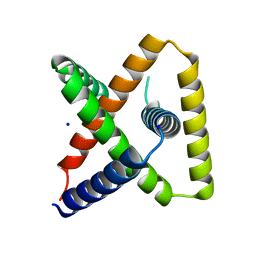 | | Designed trefoil knot protein, variant 3 | | Descriptor: | Designed trefoil knot protein, variant 3, SODIUM ION | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
1MOW
 
 | | E-DreI | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | Authors: | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2002-09-10 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
1Q33
 
 | | Crystal structure of human ADP-ribose pyrophosphatase NUDT9 | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, beta-D-glucopyranose | | Authors: | Shen, B.W, Perraud, A.L, Scharenberg, A, Stoddard, B.L. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure and Mutational Analysis of Human NUDT9
J.Mol.Biol., 332, 2003
|
|
1N3F
 
 | | Crystal structure of I-CreI bound to a palindromic DNA sequence II (palindrome of right side of wildtype DNA target sequence) | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G-3'), CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1N3E
 
 | | Crystal structure of I-CreI bound to a palindromic DNA sequence I (palindrome of left side of wildtype DNA target sequence) | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*C)-3', 5'-D(P*GP*AP*CP*GP*TP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1OX7
 
 | |
1M5X
 
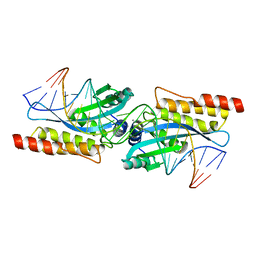 | | Crystal structure of the homing endonuclease I-MsoI bound to its DNA substrate | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-07-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
3HYI
 
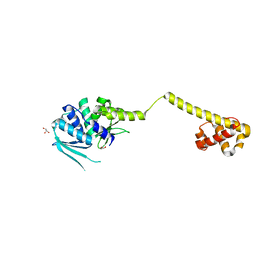 | | Crystal structure of full-length DUF199/WhiA from Thermatoga maritima | | Descriptor: | GLYCEROL, Protein DUF199/WhiA, SODIUM ION | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Structure, 17, 2009
|
|
3I1C
 
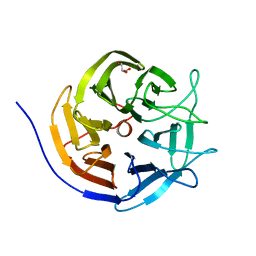 | |
3HYJ
 
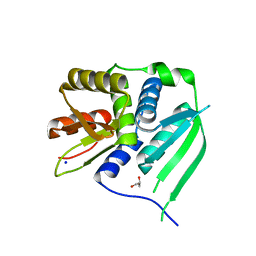 | | Crystal structure of the N-terminal LAGLIDADG domain of DUF199/WhiA | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DUF199/WhiA, ... | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
Structure, 17, 2009
|
|
5TGQ
 
 | | Restriction-modification system Type II R.SwaI, DNA free | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Shen, B.W, stoddard, B.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | DNA recognition by the SwaI restriction endonuclease involves unusual distortion of an 8 base pair A:T-rich target.
Nucleic Acids Res., 45, 2017
|
|
1CZ0
 
 | | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI/DNA COMPLEX LACKING CATALYTIC METAL ION | | Descriptor: | DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SODIUM ION, ... | | Authors: | Galburt, E.A, Chevalier, B, Jurica, M.S, Flick, K.E, Stoddard, B.L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel endonuclease mechanism directly visualized for I-PpoI.
Nat.Struct.Biol., 6, 1999
|
|
1EVW
 
 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
5TGX
 
 | |
