7XST
 
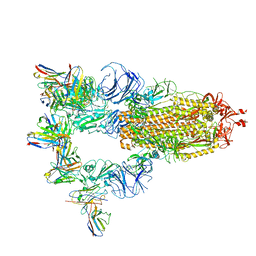 | |
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XS8
 
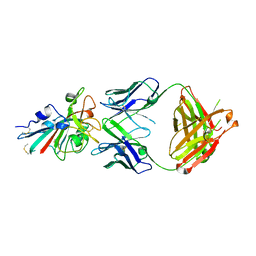 | |
7XSA
 
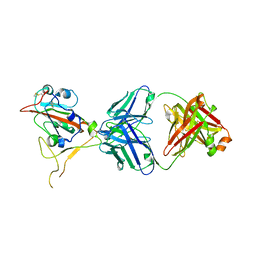 | |
7XSC
 
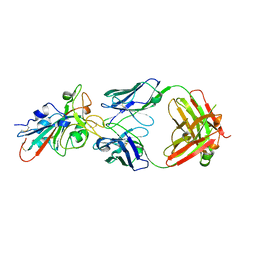 | |
7XSB
 
 | |
4YC9
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i) | | Descriptor: | GLYCEROL, N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-3,4-dimethoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4BBF
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | (2R)-N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]pyrrolidine-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BBE
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]ethanamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4YAB
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | Descriptor: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAD
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzene-1-sulfonamide (3b) | | Descriptor: | 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzenesulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBM
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (7b) | | Descriptor: | GLYCEROL, N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-3,4-dimethoxybenzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBS
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7g) | | Descriptor: | DIMETHYL SULFOXIDE, N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAT
 
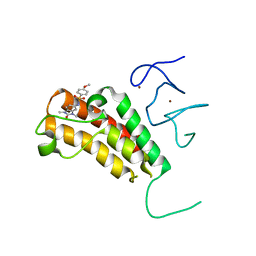 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b) | | Descriptor: | N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-4-methoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ZINC ION | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBT
 
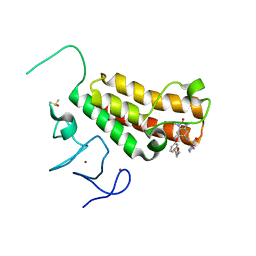 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | Descriptor: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAX
 
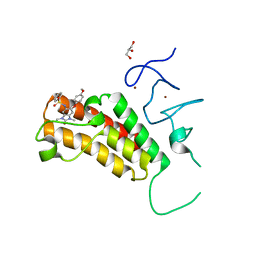 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | Descriptor: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
6WXK
 
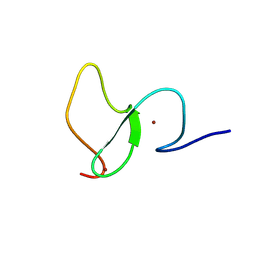 | | PHF23 PHD Domain Apo | | Descriptor: | PHD finger protein 23, ZINC ION | | Authors: | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
6WW3
 
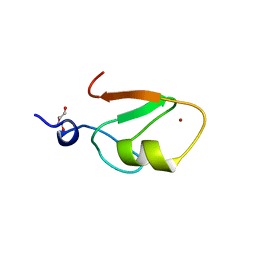 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
6KVG
 
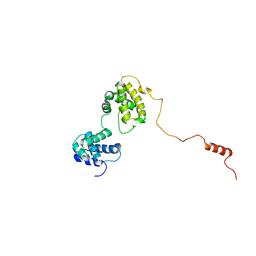 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
6WW4
 
 | | Crystal structure of HERC2 ZZ domain in complex with histone H3 tail | | Descriptor: | GLYCEROL, Histone H3.1,E3 ubiquitin-protein ligase HERC2, ZINC ION | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
9EBM
 
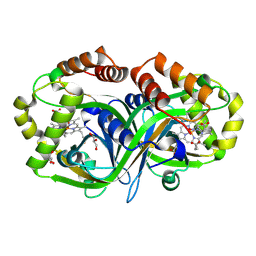 | |
9EBK
 
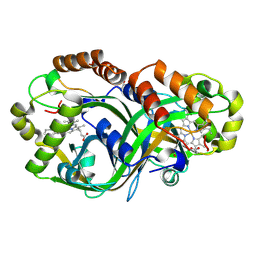 | | Piperazate synthase (PipS) in complex with haem | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Piperazate synthase | | Authors: | Higgins, M.A, Ryan, K.S. | | Deposit date: | 2024-11-12 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.08006716 Å) | | Cite: | Structure and mechanism of haem-dependent nitrogen-nitrogen bond formation in piperazate synthase
Nat Catal, 2025
|
|
7XED
 
 | | Crystal Structure of OsCIE1-Ubox and OsUBC8 complex | | Descriptor: | U-box domain-containing protein 12, UBC core domain-containing protein | | Authors: | Zhang, Y, Yu, C.Z. | | Deposit date: | 2022-03-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
9DK9
 
 | |
9DKB
 
 | |
