4WUB
 
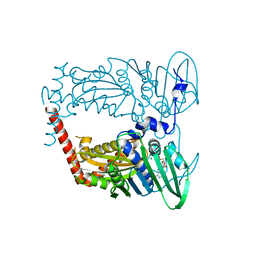 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7PTF
 
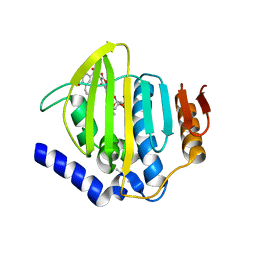 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PTG
 
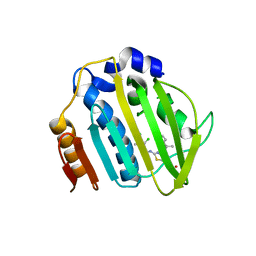 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQI
 
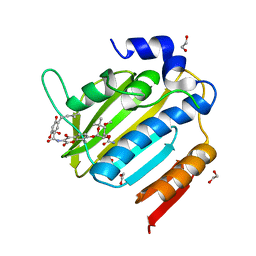 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQM
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2M
 
 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2X
 
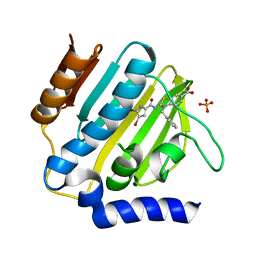 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | Descriptor: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
7P2N
 
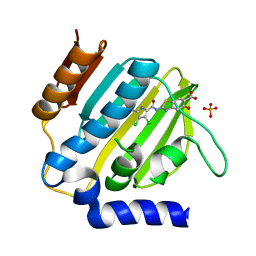 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
7QYR
 
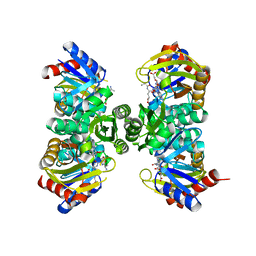 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
7QYS
 
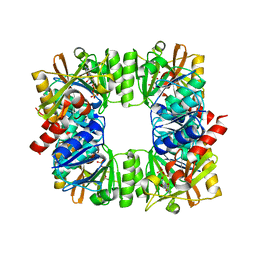 | | Crystal structure of RimK from Pseudomonas syringae DC3000 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
2WK1
 
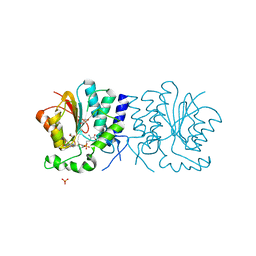 | | Structure of the O-methyltransferase NovP | | Descriptor: | 1,2-ETHANEDIOL, NOVP, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gomez Garcia, I, Stevenson, C.E.M, Uson, I, Freel Meyers, C.L, Walsh, C.T, Lawson, D.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Novobiocin Biosynthetic Enzyme Novp: The First Representative Structure for the Tylf O-Methyltransferase Superfamily.
J.Mol.Biol., 395, 2010
|
|
8A3C
 
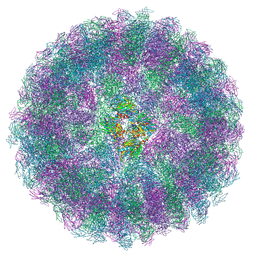 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
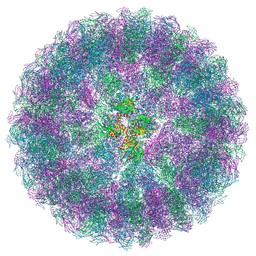 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AC6
 
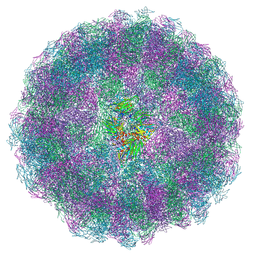 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AAY
 
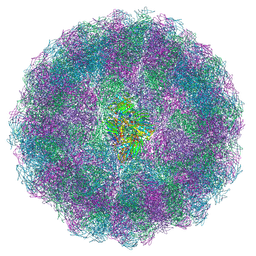 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): small class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8A41
 
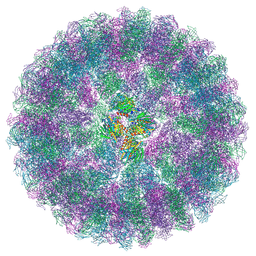 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8ACH
 
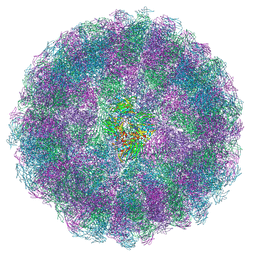 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8A3N
 
 | | Geissoschizine synthase from Catharanthus roseus - binary complex with NADP+ | | Descriptor: | Geissoschizine synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Langley, C, Tatsis, E, Hong, B, Nakamura, Y, Kamileen, M.O, Paetz, C, Stevenson, C.E.M, Basquin, J, Lawson, D.M, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
2C0Z
 
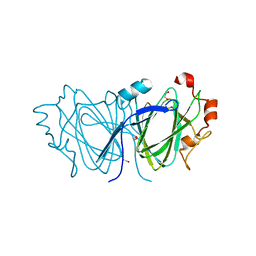 | | The 1.6 A resolution crystal structure of NovW: a 4-keto-6-deoxy sugar epimerase from the novobiocin biosynthetic gene cluster of Streptomyces spheroides | | Descriptor: | 1,2-ETHANEDIOL, NOVW, SULFATE ION | | Authors: | Jakimowicz, P, Tello, M, Freel-Meyers, C.L, Walsh, C.T, Buttner, M.J, Field, R.A, Lawson, D.M. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Resolution Crystal Structure of Novw: A 4-Keto-6-Deoxy Sugar Epimerase from the Novobiocin Biosynthetic Gene Cluster of Streptomyces Spheroides
Proteins, 63, 2006
|
|
2CJJ
 
 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
