4JMG
 
 | |
4JEG
 
 | | Crystal Structure of Monobody CS1/SHP2 C-SH2 Domain Complex | | Descriptor: | Monobody CS1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5E08
 
 | | Specific Recognition of a Single-stranded RNA Sequence by an Engineered Synthetic Antibody Fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, RNA | | Authors: | Huang, H, Qin, D, Li, N, Shao, Y, Staley, J.P, Kossiakoff, A.A, Koide, S, Piccirilli, J.A. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment.
J.Mol.Biol., 428, 2016
|
|
5DC0
 
 | |
5NKQ
 
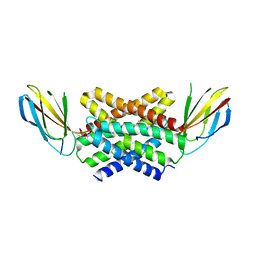 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | FLUORIDE ION, Monobody, Putative fluoride ion transporter CrcB, ... | | Authors: | Stockbridge, R, Miller, C, Newstead, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of a double-barrelled fluoride ion channel.
Nature, 525, 2015
|
|
5CWT
 
 | |
4JQ5
 
 | |
4JNU
 
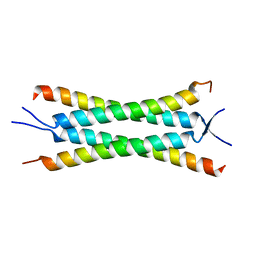 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWS
 
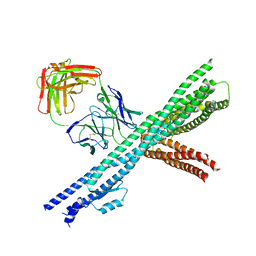 | | Crystal structure of the intact Chaetomium thermophilum Nsp1-Nup49-Nup57 channel nucleoporin heterotrimer bound to its Nic96 nuclear pore complex attachment site | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP49, ... | | Authors: | Bley, C.J, Petrovic, S, Paduch, M, Lu, V, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JO7
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex with 2:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-17 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JO9
 
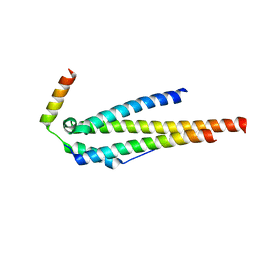 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
6C83
 
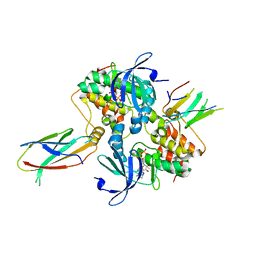 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | Descriptor: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6BF8
 
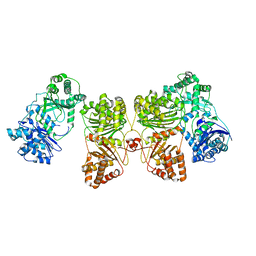 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Y
 
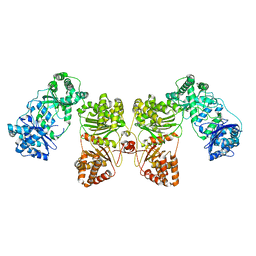 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
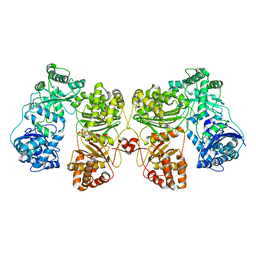 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFC
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | Descriptor: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6UX8
 
 | |
6WU1
 
 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU2
 
 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WK8
 
 | |
