6P7W
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7V
 
 | | Structure of the K. lactis CBF3 core | | Descriptor: | Cep3, Ctf13, Skp1 | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6PP7
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POD
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP5
 
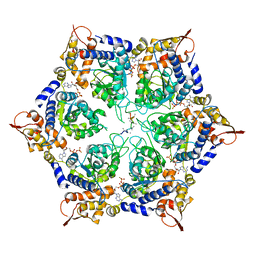 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6P7X
 
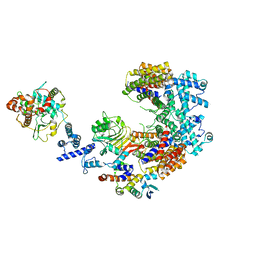 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6PO3
 
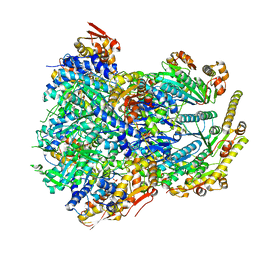 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PPE
 
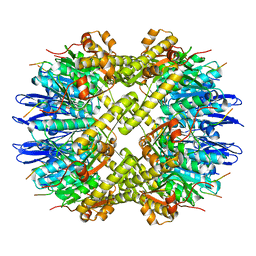 | | ClpP and ClpX IGF loop in ClpX-ClpP complex with D7 symmetry | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POS
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO1
 
 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP8
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6Q1G
 
 | |
6PWU
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-07-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6Q1E
 
 | |
6Q0H
 
 | |
6Q18
 
 | |
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6Q19
 
 | |
6Q1J
 
 | |
6Q1A
 
 | |
1VPN
 
 | | UNASSEMBLED POLYOMAVIRUS VP1 PENTAMER | | Descriptor: | POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
6Q0L
 
 | |
6Q0I
 
 | |
