1ZA8
 
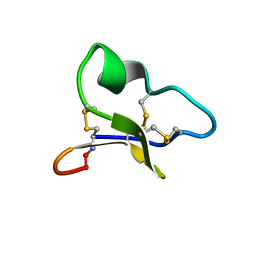 | | NMR solution structure of a leaf-specific-expressed cyclotide vhl-1 | | Descriptor: | vhl-1 | | Authors: | Chen, B, Colgrave, M.L, Daly, N.L, Rosengren, K.J, Gustafson, K.R, Craik, D.J. | | Deposit date: | 2005-04-05 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide.
J.Biol.Chem., 280, 2005
|
|
1ORX
 
 | | Solution Structure of the acyclic permutant des-(24-28)-kalata B1. | | Descriptor: | kalata B1 | | Authors: | Barry, D.G, Daly, N.L, Clark, R.J, Sando, L, Craik, D.J. | | Deposit date: | 2003-03-17 | | Release date: | 2003-06-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity
Biochemistry, 42, 2003
|
|
1T9E
 
 | |
1Q71
 
 | | The structure of microcin J25 is a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Clark, R, Daly, N.L, Goransson, U, Jones, A, Craik, D.J. | | Deposit date: | 2003-08-14 | | Release date: | 2003-12-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone.
J.Am.Chem.Soc., 125, 2003
|
|
2AB9
 
 | |
1Q8L
 
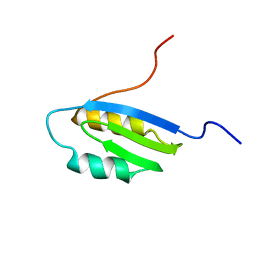 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
1XGC
 
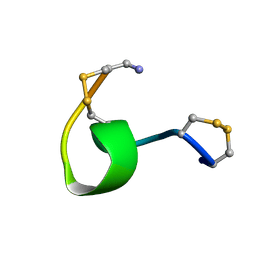 | |
1XGB
 
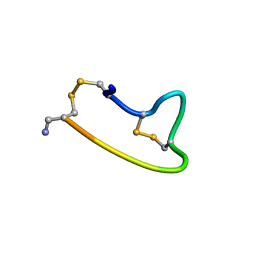 | |
1XGA
 
 | |
1VB8
 
 | |
1S7P
 
 | | Solution structure of thermolysin digested microcin J25 | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Blond, A, Afonso, C, Tabet, J.C, Rebuffat, S, Craik, D.J. | | Deposit date: | 2004-01-30 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Biochemistry, 43, 2004
|
|
1MII
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN MII | | Descriptor: | PROTEIN (ALPHA CONOTOXIN MII) | | Authors: | Hill, J.M, Oomen, C.J, Miranda, L.P, Bingham, J.P, Alewood, P.F, Craik, D.J. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity.
Biochemistry, 37, 1998
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2H8B
 
 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
8F2F
 
 | |
8FLP
 
 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
7JNN
 
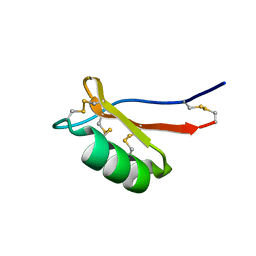 | |
7JN6
 
 | |
7KPD
 
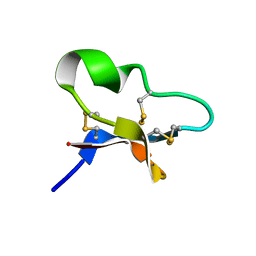 | |
7K7X
 
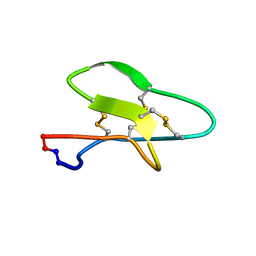 | |
7LHC
 
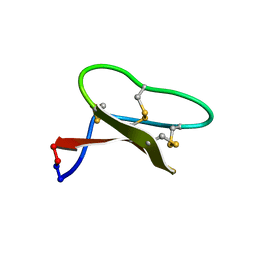 | |
6BL9
 
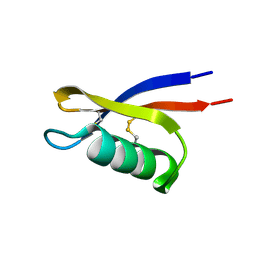 | |
6CT4
 
 | | TFE-induced NMR structure of an antimicrobial peptide (EcDBS1R5) derived from a mercury transporter protein (MerP - Escherichia coli) | | Descriptor: | EcDBS1R5 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Computationally Designed Peptide Derived from Escherichia coli as a Potential Drug Template for Antibacterial and Antibiofilm Therapies.
ACS Infect Dis, 4, 2018
|
|
6CT1
 
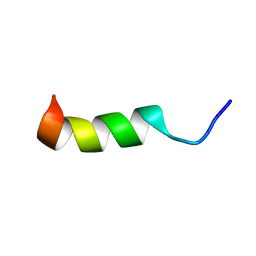 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R7) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R7 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
6CSZ
 
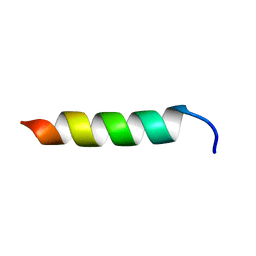 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R3) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R3 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
