4A9G
 
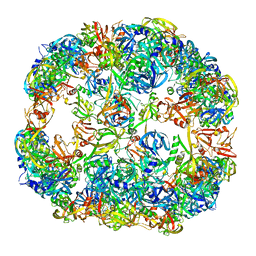 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8B
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8C
 
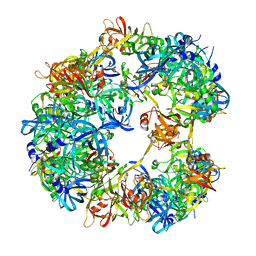 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
5D4W
 
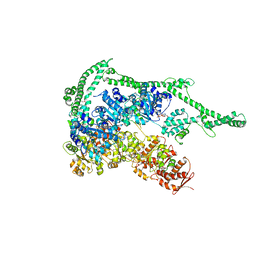 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
2HS6
 
 | |
2HS8
 
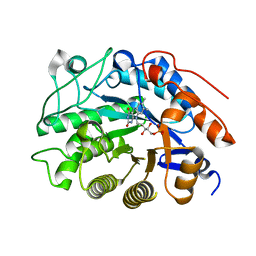 | |
2HSA
 
 | | Crystal structure of 12-oxophytodienoate reductase 3 (OPR3) from tomato | | Descriptor: | 12-oxophytodienoate reductase 3, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Clausen, T, Huber, R. | | Deposit date: | 2006-07-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1HZ4
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION FACTOR MALT DOMAIN III | | Descriptor: | BENZOIC ACID, GLYCEROL, MALT REGULATORY PROTEIN, ... | | Authors: | Steegborn, C, Danot, O, Clausen, T, Huber, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization.
Structure, 9, 2001
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
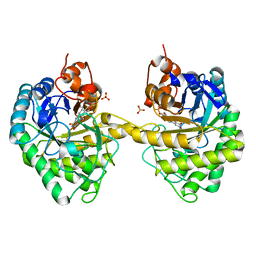
1I43
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR PPCA | | Descriptor: | 3-(PHOSPHONOMETHYL)PYRIDINE-2-CARBOXYLIC ACID, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I41
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR APPA | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-IMINO]-5-PHOSPHONO-PENT-3-ENOIC ACID, CYSTATHIONINE GAMMA-SYNTHASE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I48
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR CTCPO | | Descriptor: | CARBOXYMETHYLTHIO-3-(3-CHLOROPHENYL)-1,2,4-OXADIAZOL, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-20 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1IBJ
 
 | | Crystal structure of cystathionine beta-lyase from Arabidopsis thaliana | | Descriptor: | CARBONATE ION, CYSTATHIONINE BETA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Breitinger, U, Clausen, T, Messerschmidt, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity
Plant Physiol., 126, 2001
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1KL7
 
 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
5JYK
 
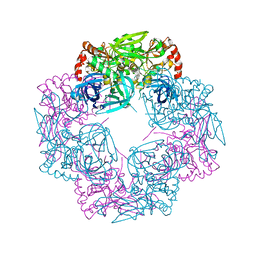 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
2ZLE
 
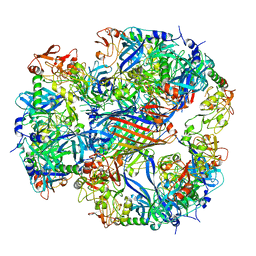 | | Cryo-EM structure of DegP12/OMP | | Descriptor: | Outer membrane protein C, Protease do | | Authors: | Schaefer, E, Saibil, H.R. | | Deposit date: | 2008-04-09 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
1GYL
 
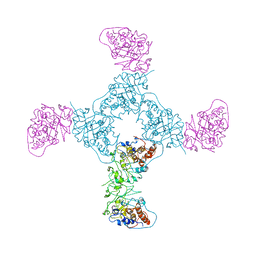 | |
1ECX
 
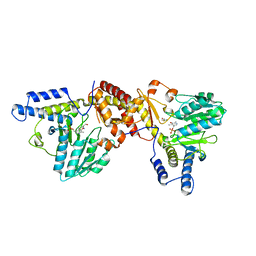 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
3OU0
 
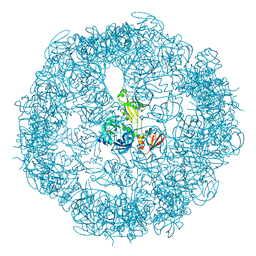 | | re-refined 3CS0 | | Descriptor: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | Authors: | Sauer, R.T, Grant, R.A, Kim, S. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
4RQZ
 
 | |
4RR0
 
 | |
