3V6K
 
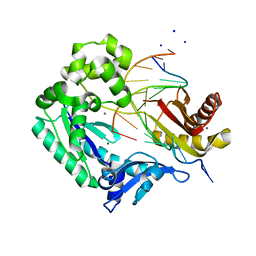 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*AP*CP*(EFG)P*GP*AP*AP*TP*CP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*GP*AP*TP*TP*CP*(2DT))-3'), ... | | Authors: | Zhao, L. | | Deposit date: | 2011-12-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8AZE
 
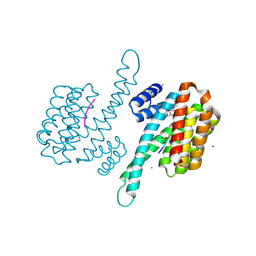 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075306) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-[(4-chlorophenyl)amino]cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, ERalpha peptide, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
4BUS
 
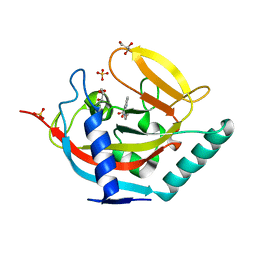 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(4-oxo-3, 4-dihydroquinazolin-2-yl)phenoxy)acetic acid | | Descriptor: | 2-[4-(4-oxidanylidene-3H-quinazolin-2-yl)phenoxy]ethanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
5XXU
 
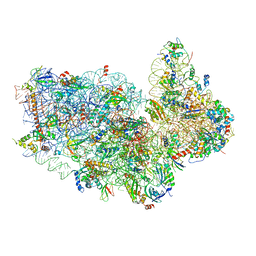 | | Small subunit of Toxoplasma gondii ribosome | | Descriptor: | 18S RNA, Ribosomal protein eL41, Ribosomal protein eS1, ... | | Authors: | LI, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
1EI2
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE RNA MAJOR GROOVE IN THE TAU EXON 10 SPLICING REGULATORY ELEMENT BY AMINOGLYCOSIDE ANTIBIOTICS | | Descriptor: | NEOMYCIN, TAU EXON 10 SRE RNA | | Authors: | Varani, L, Spillantini, M.G, Goedert, M, Varani, G. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics.
Nucleic Acids Res., 28, 2000
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4CBT
 
 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, ZINC ION | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
3V5A
 
 | | Crystal Structure of C-lobe of Bovine Lactoferrin Complexed with Gamma Amino Butyric Acid at 1.44 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-TERMINAL PEPTIDE OF LACTOTRANSFERRIN, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure of C-lobe of Bovine Lactoferrin Complexed with Gamma Amino Butyric Acid at 1.44 A Resolution
To be Published
|
|
4BUF
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4- acetylphenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-(4-acetylphenyl)-3,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
8TWL
 
 | |
4BUW
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(2-oxo-1, 3-oxazolidin-3-yl)phenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-[4-(2-OXO-1,3-OXAZOLIDIN-3-YL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
8AXU
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075297) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-(4-chloranylphenoxy)cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
5MKU
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
8TUE
 
 | |
5MMC
 
 | |
8TXU
 
 | | Fab 3864-10 in complex with influenza HA H3-SING16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 3864-10 Heavy Chain, Fab 3864-10 Light Chain, ... | | Authors: | Morano, N.C, Shapiro, L. | | Deposit date: | 2023-08-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Fab 3864-6 in complex with influenza HA H3-VIC11
To Be Published
|
|
8TWM
 
 | |
8TX3
 
 | | Fab 3864-6 in complex with influenza HA H3-VIC11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 3864-6 Heavy Chain, Fab 3864-6 Light Chain, ... | | Authors: | Morano, N.C, Shapiro, L. | | Deposit date: | 2023-08-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Fab 3864-6 in complex with influenza HA H3-VIC11
To Be Published
|
|
5Y4R
 
 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
1OAJ
 
 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castelli, S, Falconi, M, Parrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-27 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
8QFA
 
 | |
5MQ9
 
 | | Crystal structure of Rae1 (YacP) from Bacillus subtilis (W164L mutant) | | Descriptor: | SULFATE ION, Uncharacterized protein YacP | | Authors: | Piton, J, Gilet, L, Pellegrini, O, Leroy, M, Figaro, S, Condon, C. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | Rae1/YacP, a new endoribonuclease involved in ribosome-dependent mRNA decay in Bacillus subtilis.
EMBO J., 36, 2017
|
|
3VCE
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 18.1 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
3VCJ
 
 | | Thaumatin by LB Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
1EZ0
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
