6CNW
 
 | | STRUCTURE OF HUMANIZED SINGLE DOMAIN ANTIBODY SD84 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, humanized antibody SD84h | | Authors: | Luo, J, Obmolova, O. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
6W4Y
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with hydantoin stabilizer | | Descriptor: | 2-[(4~{R})-4-(2-methylpropyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]-~{N}-[4-(trifluoromethyl)phenyl]ethanamide, GLYCEROL, JTO light chain, ... | | Authors: | Yan, N.L, Morgan, G.J, Kelly, J.W. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for the stabilization of amyloidogenic immunoglobulin light chains by hydantoins.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6CNV
 
 | | INFLUENZA B/BRISBANE HEMAGGLUTININ FAB CR9115 SD84H COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR9114 Fab heavy chain, ... | | Authors: | Luo, J, Obmolova, G. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
3F1Z
 
 | |
3FMS
 
 | | Crystal structure of TM0439, a GntR transcriptional regulator | | Descriptor: | ACETATE ION, NICKEL (II) ION, Transcriptional regulator, ... | | Authors: | Zheng, M, Cooper, D.R, Yu, M, Hung, L.-W, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn2+-binding FCD domains.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3E0F
 
 | |
3DUE
 
 | |
3CGH
 
 | |
3D00
 
 | |
3CM1
 
 | |
3FKQ
 
 | |
3DL1
 
 | |
3DCX
 
 | |
3G23
 
 | |
3G0T
 
 | |
3DEE
 
 | |
3EQX
 
 | |
3ETN
 
 | |
3H41
 
 | |
1F58
 
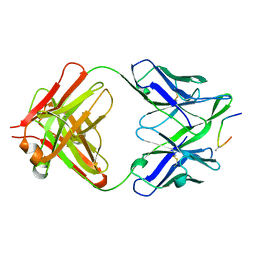 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
8CYA
 
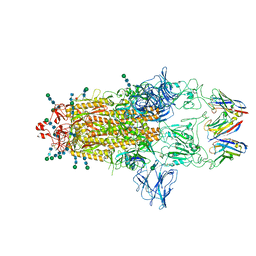 | | SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
8CXQ
 
 | |
8CY6
 
 | |
8CYC
 
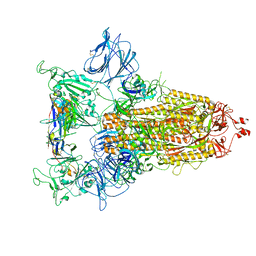 | |
8CYJ
 
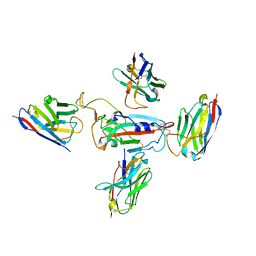 | | RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25 | | Descriptor: | Spike glycoprotein, pan-sarbecovirus nanobody 1-25, pan-sarbecovirus nanobody 2-10, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
