4OD1
 
 | | Crystal structure of human Fab CAP256-VRC26.03, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.03 heavy chain, CAP256-VRC26.03 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4PY8
 
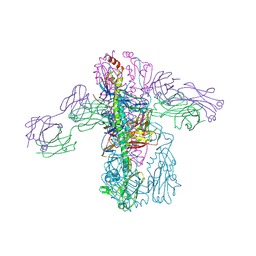 | | Crystal structure of Fab 3.1 in complex with the 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
4QDG
 
 | |
4Q5K
 
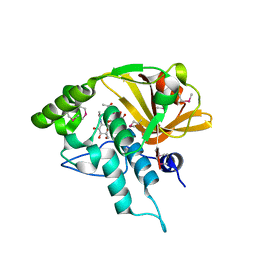 | | Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.30 A resolution | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, SODIUM ION, Uncharacterized protein | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family.
Structure, 22, 2014
|
|
4Q98
 
 | |
4QB7
 
 | |
4Q68
 
 | |
4R03
 
 | |
4R8O
 
 | |
3L4B
 
 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | ADENOSINE MONOPHOSPHATE, TrkA K+ Channel protein TM1088A, TrkA K+ Channel protein TM1088B, ... | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
4OD3
 
 | | Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CAP256-VRC26.07 heavy chain, CAP256-VRC26.07 light chain, ... | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ORG
 
 | | Crystal structure of human Fab CAP256-VRC26.04, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.04 heavy chain, CAP256-VRC26.04 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.121 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4RDB
 
 | |
3JXO
 
 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | TrkA-N domain protein | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-09-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
4OCR
 
 | | Crystal structure of human Fab CAP256-VRC26.01, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.01 heavy chain, CAP256-VRC26.01 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4RBO
 
 | |
4PY7
 
 | | Crystal Structure of Fab 3.1 | | Descriptor: | antibody 3.1 heavy chain, antibody 3.1 light chain | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
1YPI
 
 | |
1PSH
 
 | |
8SIT
 
 | |
8SIS
 
 | |
8SIR
 
 | |
8SIQ
 
 | |
1FO0
 
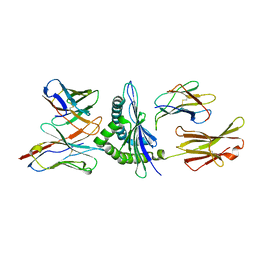 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
2FNA
 
 | |
