1S01
 
 | |
4HH4
 
 | | Structure of the CcbJ Methyltransferase from Streptomyces caelestis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CcbJ, GLYCEROL, ... | | Authors: | Bauer, J.A, Ondrovicova, G, Kutejova, E, Janata, J. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1B9R
 
 | | TERPREDOXIN FROM PSEUDOMONAS SP. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (TERPREDOXIN) | | Authors: | Mo, H, Pochapsky, S.S, Pochapsky, T.C. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas.
Biochemistry, 38, 1999
|
|
1BEV
 
 | | BOVINE ENTEROVIRUS VG-5-27 | | Descriptor: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | Authors: | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | Deposit date: | 1996-04-03 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
1BC6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1Z09
 
 | | Solution structure of km23 | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Ilangovan, U, Ding, W, Mulder, K, Hinck, A.P, Zuniga, J, Trbovich, J.A, Demeler, B, Groppe, J.C. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Homodimeric Dynein Light Chain km23.
J.Mol.Biol., 352 (2), 2005
|
|
1BW0
 
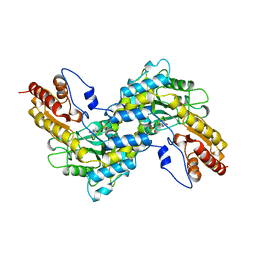 | | CRYSTAL STRUCTURE OF TYROSINE AMINOTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | PROTEIN (TYROSINE AMINOTRANSFERASE) | | Authors: | Blankenfeldt, W, Montemartini, M, Hunter, G.R, Kalisz, H.M, Nowicki, C, Hecht, H.J. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
Protein Sci., 8, 1999
|
|
1BJK
 
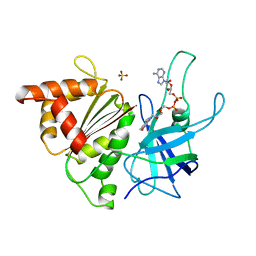 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
1BMG
 
 | |
1BJ1
 
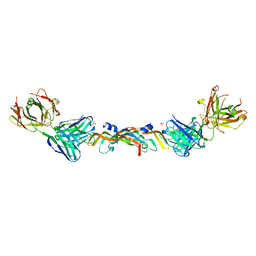 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH A NEUTRALIZING ANTIBODY | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Muller, Y.A, Christinger, H.W, De Vos, A.M. | | Deposit date: | 1998-06-30 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | VEGF and the Fab fragment of a humanized neutralizing antibody: crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface.
Structure, 6, 1998
|
|
1CRH
 
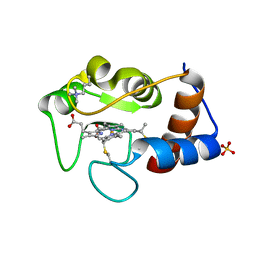 | |
1D1V
 
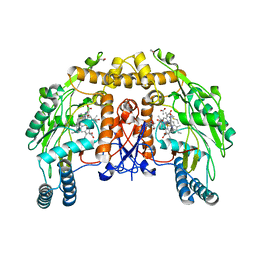 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH S-ETHYL-N-PHENYL-ISOTHIOUREA (H4B BOUND) | | Descriptor: | 2-ETHYL-1-PHENYL-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
1APB
 
 | |
220L
 
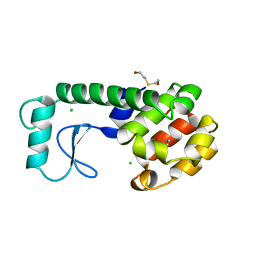 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
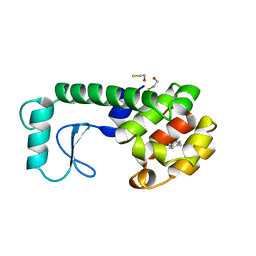 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1ZRV
 
 | | solution structure of spinigerin in H20/TFE 50% | | Descriptor: | spinigerin | | Authors: | Landon, C, Meudal, H, Boulanger, N, Bulet, P, Vovelle, F. | | Deposit date: | 2005-05-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation.
Biopolymers, 81, 2006
|
|
4J9E
 
 | |
1C6X
 
 | |
1C6Y
 
 | |
1CRI
 
 | |
4J9D
 
 | |
1ZRX
 
 | | solution structure of stomoxyn in H20/TFE 50% | | Descriptor: | stomoxyn | | Authors: | Landon, C, Meudal, H, Boulanger, N, Bulet, P, Vovelle, F. | | Deposit date: | 2005-05-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation.
Biopolymers, 81, 2006
|
|
1ZRW
 
 | | solution structure of spinigerin in H20/TFE 10% | | Descriptor: | spinigerin | | Authors: | Landon, C, Meudal, H, Boulanger, N, Bulet, P, Vovelle, F. | | Deposit date: | 2005-05-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation.
Biopolymers, 81, 2006
|
|
