7BEF
 
 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7QU5
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTY
 
 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | 1-(furan-2-ylmethyl)-3-(2-methylphenyl)thiourea, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU3
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU0
 
 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6IDA
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID8
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTL ( A138S, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
3SQ1
 
 | | RB69 DNA Polymerase Ternary Complex with dUpCpp Opposite dA | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
6ID3
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGPL ( A138S, V186G) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-08 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDD
 
 | | Crystal structure of H7 hemagglutinin mutant SH1-AVPL ( S138A, G186V, T221P, Q226L) from the influenza virus A/Shanghai/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
6ICX
 
 | |
6ID9
 
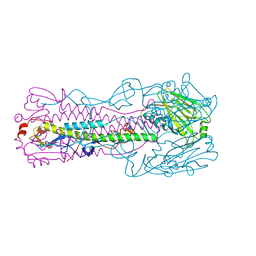 | | Crystal structure of H7 hemagglutinin mutant H7-SGTL ( A138S, V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IF2
 
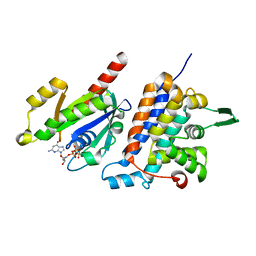 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
5YS2
 
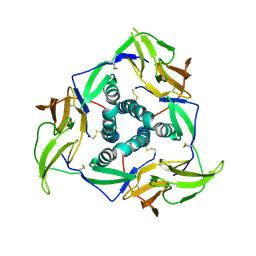 | |
5NGD
 
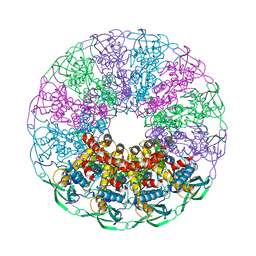 | | Crystal structure of the V325G mutant of the bacteriophage G20C portal protein | | Descriptor: | Portal protein | | Authors: | Pascoa, T, Jenkins, H.T, Chechik, M, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization
Acs Nano, 11, 2017
|
|
6ICY
 
 | | Crystal structure of H7 hemagglutinin mutant H7-AGTL ( V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-07 | | Release date: | 2019-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDB
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
5YSL
 
 | | Crystal structure of antibody 1H1 Fab | | Descriptor: | 1H1 heavy chain, 1H1 light chain | | Authors: | Hu, X.L, Yang, F.L. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design.
PLoS Pathog., 13, 2017
|
|
3SPY
 
 | | RB69 DNA Polymerase(L415A/L561A/S565G/Y567A) Ternary Complex with dUpCpp Opposite dA | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
3CGL
 
 | |
6IZO
 
 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
8SXG
 
 | |
8SXE
 
 | |
8SXH
 
 | |
