2O7L
 
 | | The open-cap conformation of GlpG | | Descriptor: | Protein glpG, nonyl beta-D-glucopyranoside | | Authors: | Ha, Y. | | Deposit date: | 2006-12-11 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-cap conformation of intramembrane protease GlpG.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8XQ3
 
 | | Structure of Nipah virus Bangladesh string G protein ectodomain tetramer bound to single-domain antibody n425 at 5.87 Angstroms overall resolution | | Descriptor: | Nipah virus Bangladesh string G protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
8XPY
 
 | | Structure of Nipah virus Malaysia string G protein ectodomain monomer bound to single-domain antibody n425 at 3.63 Angstroms overall resolution | | Descriptor: | Glycoprotein G, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
8XPS
 
 | | Structure of Nipah virus Bangladesh string G protein ectodomain monomer bound to single-domain antibody n425 at 3.22 Angstroms overall resolution | | Descriptor: | Nipah virus Bangladesh string G protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
8TXC
 
 | |
8TX1
 
 | |
8TXB
 
 | |
5TRL
 
 | |
5TRM
 
 | |
3TSY
 
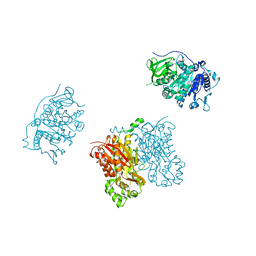 | | 4-Coumaroyl-CoA Ligase::Stilbene Synthase fusion protein | | Descriptor: | Fusion Protein 4-coumarate--CoA ligase 1, Resveratrol synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Kinetic Analysis of the Unnatural Fusion Protein 4-Coumaroyl-CoA Ligase::Stilbene Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
9IJB
 
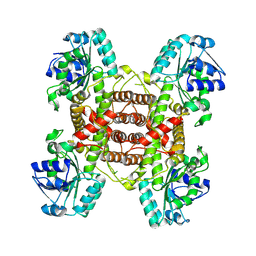 | | Crystal structure and function analysis of a highly potential drug target 6-phosphogluconate dehydrogenase in Mycobacterium tuberculosis | | Descriptor: | 6-phosphogluconate dehydrogenase, NAD(+)-dependent, decarboxylating | | Authors: | Wang, Y.Z, Ren, X.Q, Li, T, Zhang, R.D. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.740405 Å) | | Cite: | Crystal structure and function analysis of 6-phosphogluconate dehydrogenase in Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 731, 2024
|
|
5XYZ
 
 | | The structure of human BTK kinase domain in complex with a covalent inhibitor | | Descriptor: | N-[3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-1H-1,2,4-triazol-3-yl)phenyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Xie, Y.T, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5XYX
 
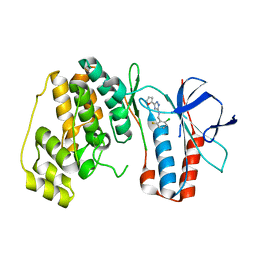 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5XYY
 
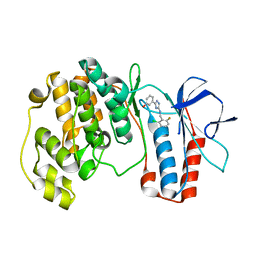 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
3A1L
 
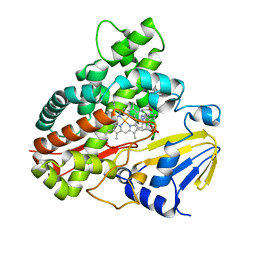 | | Crystal Structure of 11,11'-Dichlorochromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 3,4-bis(7-chloro-1H-indol-3-yl)-1H-pyrrole-2,5-dicarboxylic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Theoretical and experimental studies of the conversion of chromopyrrolic acid to an antitumor derivative by cytochrome P450 StaP: the catalytic role of water molecules
J.Am.Chem.Soc., 131, 2009
|
|
8Z9Z
 
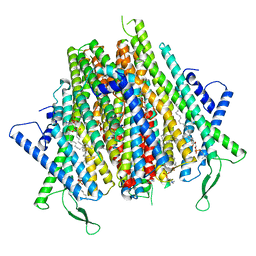 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
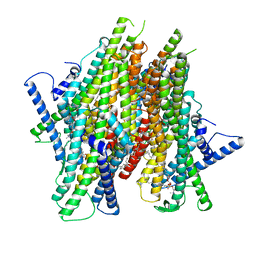 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8ZPV
 
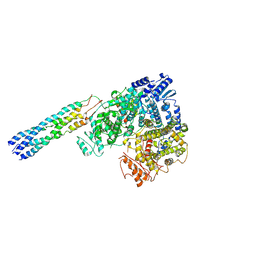 | | Nipah virus polymerase complex | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Wang, Y.R, Zhang, H.Q. | | Deposit date: | 2024-05-31 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of Nipah virus RNA polymerase complex.
Sci Adv, 10, 2024
|
|
6AE8
 
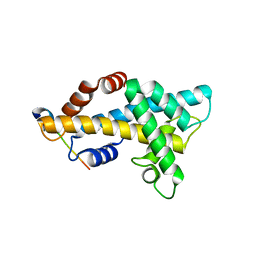 | | Structure insight into histone chaperone Chz1-mediated H2A.Z recognition and replacement | | Descriptor: | BICINE, Histone H2A.Z-specific chaperone CHZ1, Histone H2B.1,Histone H2A.Z | | Authors: | Wang, Y.Y, Shan, S, Zhou, Z. | | Deposit date: | 2018-08-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement.
Plos Biol., 17, 2019
|
|
7VGG
 
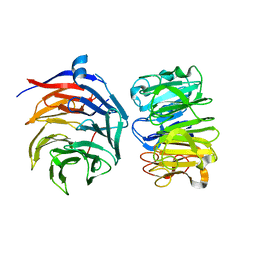 | | Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1 | | Descriptor: | E3 ubiquitin-protein ligase COP1, Ultraviolet-B receptor UVR8 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into UV-B-activated UVR8 bound to COP1.
Sci Adv, 8, 2022
|
|
7WK0
 
 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WOP
 
 | | The local refined map of Omicron spike with bispecific antibody FD01 | | Descriptor: | 16L9, GW01, Spike protein S1 | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOR
 
 | | The state 2 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
