3D79
 
 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
5C5O
 
 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
5C5N
 
 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
7Y3V
 
 | | Crystal structure of CdpNPT in complex with harmane | | Descriptor: | 1-methyl-9H-pyrido[3,4-b]indole, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-06-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic potential of a fungal indole prenyltransferase toward beta-carbolines, harmine and harman, and their prenylation effects on antibacterial activity.
J.Biosci.Bioeng., 134, 2022
|
|
7XVJ
 
 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
7W6F
 
 | | Polyketide cyclase OAC-F24I mutant from Cannabis sativa in complex with 6-nonylresorcylic acid | | Descriptor: | 2-nonyl-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, Olivetolic acid cyclase | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6G
 
 | | TKS-L190G mutant from Cannabis sativa in complex with lauroyl-CoA | | Descriptor: | 3,5,7-trioxododecanoyl-CoA synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6E
 
 | |
7W6D
 
 | |
7BMI
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-fluoropyridine-2,4-dicarboxylic acid, and factor X substrate peptide fragment (39mer-4Ser) | | Descriptor: | 3-fluoranylpyridine-2,4-dicarboxylic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Fluorinated derivatives of pyridine-2,4-dicarboxylate are potent inhibitors of human 2-oxoglutarate dependent oxygenases
J Fluor Chem, 247, 2021
|
|
7BMJ
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 5-fluoropyridine-2,4-dicarboxylic acid, and factor X substrate peptide fragment (39mer-4Ser) | | Descriptor: | 5-fluoranylpyridine-2,4-dicarboxylic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorinated derivatives of pyridine-2,4-dicarboxylate are potent inhibitors of human 2-oxoglutarate dependent oxygenases
J Fluor Chem, 247, 2021
|
|
7E6J
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H725A in complex with Factor X peptide fragment (39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Nakashima, Y, Brasnett, A, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Oxygenase Variants Employing a Single Protein Fe II Ligand Are Catalytically Active.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DYV
 
 | | Human JMJD5 in complex with MN and 5-(benzylamino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-(benzylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYW
 
 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYX
 
 | | Human JMJD5 in complex with MN and 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYU
 
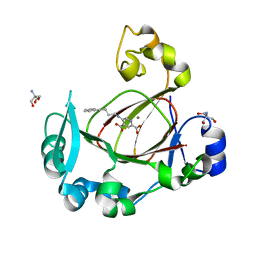 | | Human JMJD5 in complex with MN and 5-((4-phenylbutyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(4-phenylbutylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
1VF9
 
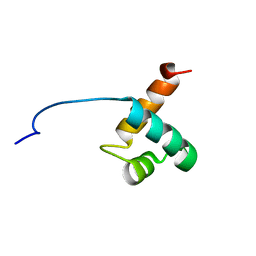 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1VFC
 
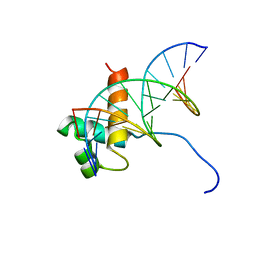 | | Solution Structure Of The DNA Complex Of Human Trf2 | | Descriptor: | Short C-rich starnd, Short G-rich strand, Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
3VWR
 
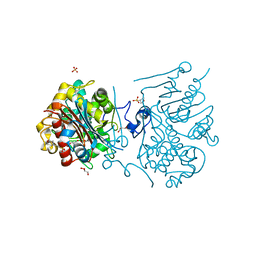 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWL
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
