1ODG
 
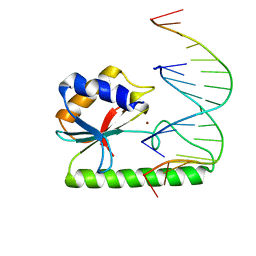 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
1AH6
 
 | |
1AH8
 
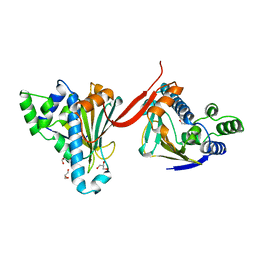 | |
1Q2Z
 
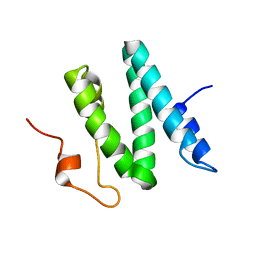 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
2CE9
 
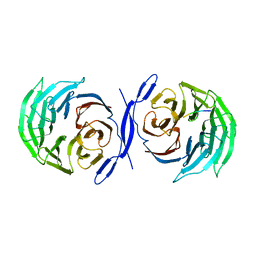 | |
2CE8
 
 | |
4AV1
 
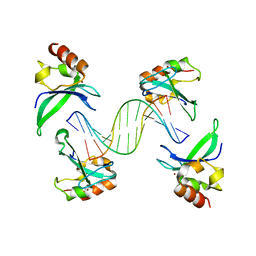 | | Crystal structure of the human PARP-1 DNA binding domain in complex with DNA | | Descriptor: | 5'-D(*AP*AP*GP*TP*GP*TP*TP*GP*CP*AP*TP*TP)-3', 5'-D(*TP*AP*AP*TP*GP*CP*AP*AP*CP*AP*CP*TP)-3', POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Ali, A.A.E, Timinszky, G, Arribas-Bosacoma, R, Kozlowski, M, Hassa, P.O, Hassler, M, Ladurner, A.G, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Zinc-Finger Domains of Parp1 Cooperate to Recognise DNA Strand-Breaks
Nat.Struct.Mol.Biol., 19, 2012
|
|
4BA9
 
 | | The structural basis for the coordination of Y-family Translesion DNA Polymerases by Rev1 | | Descriptor: | DNA POLYMERASE KAPPA, DNA REPAIR PROTEIN REV1, MAGNESIUM ION, ... | | Authors: | Grummitt, C.G, Kilkenny, M.L, Frey, A, Roe, S.M, Oliver, A.W, Sale, J.E, Pearl, L.H. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Structural Basis for the Coordination of Y- Family Translesion DNA Polymerases by Rev1
To be Published
|
|
4B7T
 
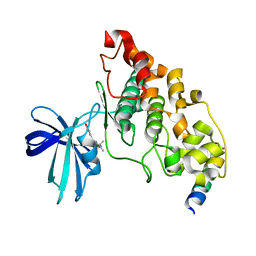 | | Glycogen Synthase Kinase 3 Beta complexed with Axin Peptide and Leucettine L4 | | Descriptor: | (5Z)-5-(1,3-benzodioxol-5-ylmethylidene)-3-methyl-2-(propan-2-ylamino)imidazol-4-one, AXIN-1, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Oberholzer, A.E, Pearl, L.H. | | Deposit date: | 2012-08-22 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Selectivity, Cocrystal Structures, and Neuroprotective Properties of Leucettines, a Family of Protein Kinase Inhibitors Derived from the Marine Sponge Alkaloid Leucettamine B.
J.Med.Chem., 55, 2012
|
|
4BB7
 
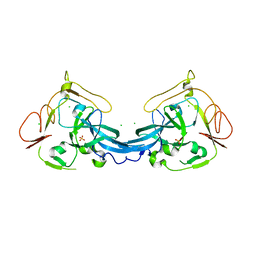 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
4BMC
 
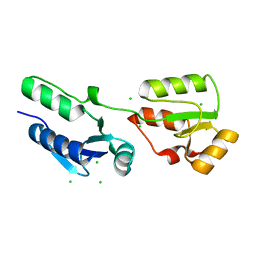 | | Crystal structure of s.pombe Rad4 BRCT1,2 | | Descriptor: | CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | Authors: | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BU1
 
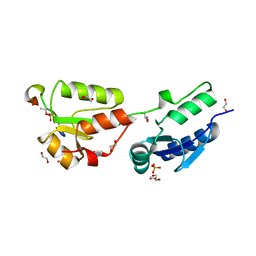 | | Crystal structure of Rad4 BRCT1,2 in complex with a Crb2 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, DNA REPAIR PROTEIN RHP9, GLYCEROL, ... | | Authors: | Qu, M, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BU0
 
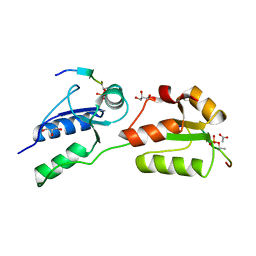 | | Crystal structure of Rad4 BRCT1,2 in complex with a Crb2 phosphopeptide | | Descriptor: | ACETATE ION, DNA REPAIR PROTEIN RHP9, GLYCEROL, ... | | Authors: | Qu, M, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
4BMD
 
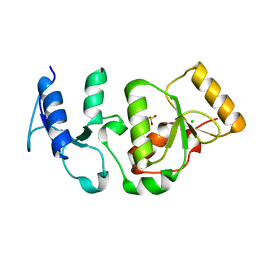 | | Crystal structure of S.pombe Rad4 BRCT3,4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, S-M CHECKPOINT CONTROL PROTEIN RAD4 | | Authors: | Meng, Q, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
1KCF
 
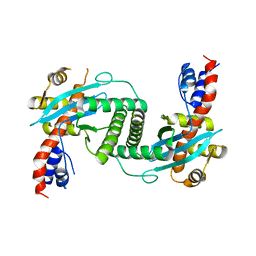 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1KO9
 
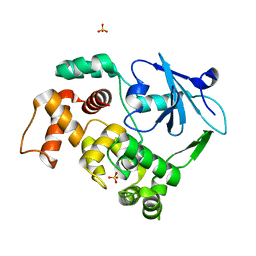 | | Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG1 | | Descriptor: | 8-oxoguanine DNA glycosylase, SULFATE ION | | Authors: | Bjoras, M, Seeberg, E, Luna, L, Pearl, L.H, Barrett, T.E. | | Deposit date: | 2001-12-20 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase.
J.Mol.Biol., 317, 2002
|
|
1MUG
 
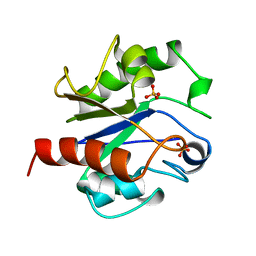 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | Descriptor: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1MWJ
 
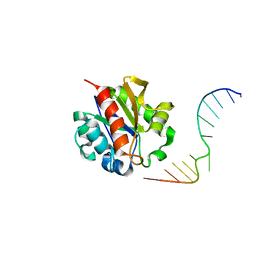 | | Crystal Structure of a MUG-DNA pseudo substrate complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*A*GP*(DU)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Scharer, O, Savva, R, Brown, T, Jiricny, J, Verdine, G.L, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of a thwarted mismatch glycosylase DNA repair complex
Embo J., 18, 1999
|
|
1MTL
 
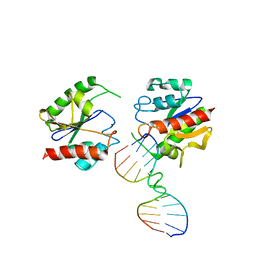 | | Non-productive MUG-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Barlow, T, Brown, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-21 | | Release date: | 2002-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Nat.Struct.Biol., 5, 1998
|
|
1MWI
 
 | | Crystal structure of a MUG-DNA product complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1OE4
 
 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DR*AP*CP*GP*GP*G)-3', GLYCEROL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1A4H
 
 | |
1GEC
 
 | | GLYCYL ENDOPEPTIDASE-COMPLEX WITH BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE COVALENTLY BOUND TO CYSTEINE 25 | | Descriptor: | BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE INHIBITOR, GLYCYL ENDOPEPTIDASE | | Authors: | Ohara, B.P, Hemmings, A.M, Buttle, D.J, Pearl, L.H. | | Deposit date: | 1995-05-25 | | Release date: | 1995-12-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
Biochemistry, 34, 1995
|
|
1GN1
 
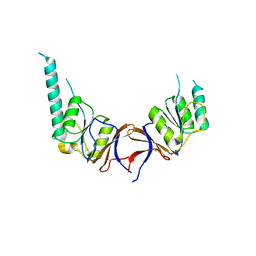 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GML
 
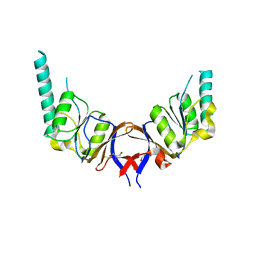 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
